A framework for oligonucleotide microarray preprocessing
- PMID: 20688976
- PMCID: PMC2944196
- DOI: 10.1093/bioinformatics/btq431
A framework for oligonucleotide microarray preprocessing
Abstract
Motivation: The availability of flexible open source software for the analysis of gene expression raw level data has greatly facilitated the development of widely used preprocessing methods for these technologies. However, the expansion of microarray applications has exposed the limitation of existing tools.
Results: We developed the oligo package to provide a more general solution that supports a wide range of applications. The package is based on the BioConductor principles of transparency, reproducibility and efficiency of development. It extends the existing tools and leverages existing code for visualization, accessing data and widely used preprocessing routines. The oligo package implements a unified paradigm for preprocessing data and interfaces with other BioConductor tools for downstream analysis. Our infrastructure is general and can be used by other BioConductor packages.
Availability: The oligo package is freely available through BioConductor, http://www.bioconductor.org.
Figures
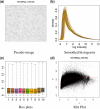


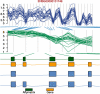
References
-
- Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
-
- Carvalho BS, et al. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. - PubMed
-
- Gautier L, et al. affy—analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20:307–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

