GeneCards Version 3: the human gene integrator
- PMID: 20689021
- PMCID: PMC2938269
- DOI: 10.1093/database/baq020
GeneCards Version 3: the human gene integrator
Abstract
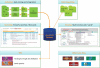
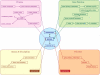
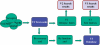
GeneCards (www.genecards.org) is a comprehensive, authoritative compendium of annotative information about human genes, widely used for nearly 15 years. Its gene-centric content is automatically mined and integrated from over 80 digital sources, resulting in a web-based deep-linked card for each of >73,000 human gene entries, encompassing the following categories: protein coding, pseudogene, RNA gene, genetic locus, cluster and uncategorized. We now introduce GeneCards Version 3, featuring a speedy and sophisticated search engine and a revamped, technologically enabling infrastructure, catering to the expanding needs of biomedical researchers. A key focus is on gene-set analyses, which leverage GeneCards' unique wealth of combinatorial annotations. These include the GeneALaCart batch query facility, which tabulates user-selected annotations for multiple genes and GeneDecks, which identifies similar genes with shared annotations, and finds set-shared annotations by descriptor enrichment analysis. Such set-centric features address a host of applications, including microarray data analysis, cross-database annotation mapping and gene-disorder associations for drug targeting. We highlight the new Version 3 database architecture, its multi-faceted search engine, and its semi-automated quality assurance system. Data enhancements include an expanded visualization of gene expression patterns in normal and cancer tissues, an integrated alternative splicing pattern display, and augmented multi-source SNPs and pathways sections. GeneCards now provides direct links to gene-related research reagents such as antibodies, recombinant proteins, DNA clones and inhibitory RNAs and features gene-related drugs and compounds lists. We also portray the GeneCards Inferred Functionality Score annotation landscape tool for scoring a gene's functional information status. Finally, we delineate examples of applications and collaborations that have benefited from the GeneCards suite. Database URL: www.genecards.org.
Figures




 between ExUns indicates an intron, while ‘·’ indicates the junction of two ExUns.
between ExUns indicates an intron, while ‘·’ indicates the junction of two ExUns.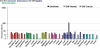




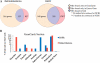
References
-
- Rebhan M., Chalifa-Caspi V., Prilusky J., et al. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;8:656–664. - PubMed
-
- Safran M., Solomon I., Shmueli O., et al. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;11:1542–1543. - PubMed
-
- HGNC. [(1 August 2010, date last accessed)]. http://www.genenames.org/
-
- Entrez gene. [(1 August 2010, date last accessed)]. http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

