Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus
- PMID: 20689043
- PMCID: PMC2930543
- DOI: 10.1073/pnas.1007071107
Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus
Abstract
The M2 proton channel from influenza A virus is an essential protein that mediates transport of protons across the viral envelope. This protein has a single transmembrane helix, which tetramerizes into the active channel. At the heart of the conduction mechanism is the exchange of protons between the His37 imidazole moieties of M2 and waters confined to the M2 bundle interior. Protons are conducted as the total charge of the four His37 side chains passes through 2(+) and 3(+) with a pK(a) near 6. A 1.65 A resolution X-ray structure of the transmembrane protein (residues 25-46), crystallized at pH 6.5, reveals a pore that is lined by alternating layers of sidechains and well-ordered water clusters, which offer a pathway for proton conduction. The His37 residues form a box-like structure, bounded on either side by water clusters with well-ordered oxygen atoms at close distance. The conformation of the protein, which is intermediate between structures previously solved at higher and lower pH, suggests a mechanism by which conformational changes might facilitate asymmetric diffusion through the channel in the presence of a proton gradient. Moreover, protons diffusing through the channel need not be localized to a single His37 imidazole, but instead may be delocalized over the entire His-box and associated water clusters. Thus, the new crystal structure provides a possible unification of the discrete site versus continuum conduction models.
Conflict of interest statement
Conflict of interest statement: R.A.L., L.H.P., W.F.D., and M.L.K. are members of the scientific advisory board of InfluMedix, a company that is developing influenza drugs.
Figures

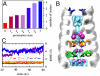
 state (light blue shading) is plotted together with the experimental one (red surfaces). The overall structure of the bundle is also presented (gray shading) and the sidechains of some of functionally relevant residues are highlighted as sticks: Val27 (blue), His37 (orange), and Trp41 (green). (C) The rmsd of the heavy atoms from the initial (X-ray) structure is plotted as a function of MD simulation time for the protonation states
state (light blue shading) is plotted together with the experimental one (red surfaces). The overall structure of the bundle is also presented (gray shading) and the sidechains of some of functionally relevant residues are highlighted as sticks: Val27 (blue), His37 (orange), and Trp41 (green). (C) The rmsd of the heavy atoms from the initial (X-ray) structure is plotted as a function of MD simulation time for the protonation states  (red),
(red),  (gold),
(gold),  (purple), and 4+ (blue), respectively.
(purple), and 4+ (blue), respectively.
References
-
- Headrick JM, et al. Spectral signatures of hydrated proton vibrations in water clusters. Science. 2005;308:1765–1769. - PubMed
-
- Shin J-W, et al. Infrared signature of structures associated with the H+(H2O)n (n = 6 to 27) Science. 2004;304:1137–1140. - PubMed
-
- Robertson WH, et al. Spectroscopic determination of the OH- solvation shell in the OH-(H2O)n clusters. Science. 2003;299:1367–1372. - PubMed
-
- Liu K, Cruzan JD, Saykally RJ. Water clusters. Science. 1996;271:929–933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

