Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization
- PMID: 20689044
- PMCID: PMC2930523
- DOI: 10.1073/pnas.1009945107
Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization
Abstract
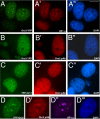
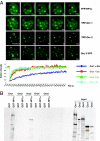
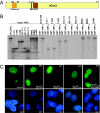
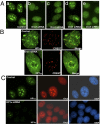
The origin recognition complex (ORC) is a DNA replication initiator protein also known to be involved in diverse cellular functions including gene silencing, sister chromatid cohesion, telomere biology, heterochromatin localization, centromere and centrosome activity, and cytokinesis. We show that, in human cells, multiple ORC subunits associate with hetereochromatin protein 1 (HP1) alpha- and HP1beta-containing heterochromatic foci. Fluorescent bleaching studies indicate that multiple subcomplexes of ORC exist at heterochromatin, with Orc1 stably associating with heterochromatin in G1 phase, whereas other ORC subunits have transient interactions throughout the cell-division cycle. Both Orc1 and Orc3 directly bind to HP1alpha, and two domains of Orc3, a coiled-coil domain and a mod-interacting region domain, can independently bind to HP1alpha; however, both are essential for in vivo localization of Orc3 to heterochromatic foci. Direct binding of both Orc1 and Orc3 to HP1 suggests that, after the degradation of Orc1 at the G1/S boundary, Orc3 facilitates assembly of ORC/HP1 proteins to chromatin. Although depletion of Orc2 and Orc3 subunits by siRNA caused loss of HP1alpha association to heterochromatin, loss of Orc1 and Orc5 caused aberrant HP1alpha distribution only to pericentric heterochromatin-surrounding nucleoli. Depletion of HP1alpha from human cells also shows loss of Orc2 binding to heterochromatin, suggesting that ORC and HP1 proteins are mutually required for each other to bind to heterochromatin. Similar to HP1alpha-depleted cells, Orc2 and Orc3 siRNA-treated cells also show loss of compaction at satellite repeats, suggesting that ORC together with HP1 proteins may be involved in organizing higher-order chromatin structure and centromere function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance.EMBO J. 2004 Jul 7;23(13):2651-63. doi: 10.1038/sj.emboj.7600255. Epub 2004 Jun 24. EMBO J. 2004. PMID: 15215892 Free PMC article.
-
Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing.Mol Biol Cell. 2001 Jun;12(6):1671-85. doi: 10.1091/mbc.12.6.1671. Mol Biol Cell. 2001. PMID: 11408576 Free PMC article.
-
Interaction between HP1alpha and replication proteins in mammalian cells.Exp Cell Res. 2006 Oct 15;312(17):3349-59. doi: 10.1016/j.yexcr.2006.07.014. Epub 2006 Jul 28. Exp Cell Res. 2006. PMID: 16950245
-
The origin recognition complex protein family.Genome Biol. 2009;10(3):214. doi: 10.1186/gb-2009-10-3-214. Epub 2009 Mar 17. Genome Biol. 2009. PMID: 19344485 Free PMC article. Review.
-
The many faces of the origin recognition complex.Curr Opin Cell Biol. 2007 Jun;19(3):337-43. doi: 10.1016/j.ceb.2007.04.007. Epub 2007 Apr 26. Curr Opin Cell Biol. 2007. PMID: 17466500 Review.
Cited by
-
DNA replication fading as proliferating cells advance in their commitment to terminal differentiation.Sci Rep. 2012;2:279. doi: 10.1038/srep00279. Epub 2012 Feb 20. Sci Rep. 2012. PMID: 22359734 Free PMC article.
-
The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin.Nat Struct Mol Biol. 2012 Mar 4;19(4):458-60. doi: 10.1038/nsmb.2244. Nat Struct Mol Biol. 2012. PMID: 22388734
-
ORChestra coordinates the replication and repair music.Bioessays. 2023 Apr;45(4):e2200229. doi: 10.1002/bies.202200229. Epub 2023 Feb 22. Bioessays. 2023. PMID: 36811379 Free PMC article.
-
Chromatin and Nuclear Dynamics in the Maintenance of Replication Fork Integrity.Front Genet. 2021 Dec 14;12:773426. doi: 10.3389/fgene.2021.773426. eCollection 2021. Front Genet. 2021. PMID: 34970302 Free PMC article. Review.
-
Heterochromatin and the molecular mechanisms of 'parent-of-origin' effects in animals.J Biosci. 2016 Dec;41(4):759-786. doi: 10.1007/s12038-016-9650-9. J Biosci. 2016. PMID: 27966495 Review.
References
-
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. - PubMed
-
- Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276:29067–29071. - PubMed
-
- Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

