ATP synthase: from single molecule to human bioenergetics
- PMID: 20689227
- PMCID: PMC3066536
- DOI: 10.2183/pjab.86.667
ATP synthase: from single molecule to human bioenergetics
Abstract
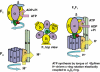
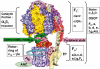
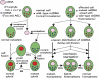
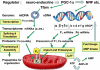
ATP synthase (F(o)F(1)) consists of an ATP-driven motor (F(1)) and a H(+)-driven motor (F(o)), which rotate in opposite directions. F(o)F(1) reconstituted into a lipid membrane is capable of ATP synthesis driven by H(+) flux. As the basic structures of F(1) (alpha(3)beta(3)gammadeltaepsilon) and F(o) (ab(2)c(10)) are ubiquitous, stable thermophilic F(o)F(1) (TF(o)F(1)) has been used to elucidate molecular mechanisms, while human F(1)F(o) (HF(1)F(o)) has been used to study biomedical significance. Among F(1)s, only thermophilic F(1) (TF(1)) can be analyzed simultaneously by reconstitution, crystallography, mutagenesis and nanotechnology for torque-driven ATP synthesis using elastic coupling mechanisms. In contrast to the single operon of TF(o)F(1), HF(o)F(1) is encoded by both nuclear DNA with introns and mitochondrial DNA. The regulatory mechanism, tissue specificity and physiopathology of HF(o)F(1) were elucidated by proteomics, RNA interference, cytoplasts and transgenic mice. The ATP synthesized daily by HF(o)F(1) is in the order of tens of kilograms, and is primarily controlled by the brain in response to fluctuations in activity.
Figures





Similar articles
-
Efficient ATP synthesis by thermophilic Bacillus FoF1-ATP synthase.FEBS J. 2011 Aug;278(15):2647-54. doi: 10.1111/j.1742-4658.2011.08191.x. Epub 2011 Jun 20. FEBS J. 2011. PMID: 21605343 Free PMC article.
-
The alpha/beta interfaces of alpha(1)beta(1), alpha(3)beta(3), and F1: domain motions and elastic energy stored during gamma rotation.J Bioenerg Biomembr. 2000 Oct;32(5):471-84. doi: 10.1023/a:1005612923995. J Bioenerg Biomembr. 2000. PMID: 15254382
-
Inhibition of ATP hydrolysis by thermoalkaliphilic F1Fo-ATP synthase is controlled by the C terminus of the epsilon subunit.J Bacteriol. 2006 Jun;188(11):3796-804. doi: 10.1128/JB.00040-06. J Bacteriol. 2006. PMID: 16707672 Free PMC article.
-
Structures and interactions of proteins involved in the coupling function of the protonmotive F(o)F(1)-ATP synthase.Curr Protein Pept Sci. 2002 Aug;3(4):451-60. doi: 10.2174/1389203023380558. Curr Protein Pept Sci. 2002. PMID: 12370007 Review.
-
IF(1): setting the pace of the F(1)F(o)-ATP synthase.Trends Biochem Sci. 2009 Jul;34(7):343-50. doi: 10.1016/j.tibs.2009.03.006. Epub 2009 Jun 24. Trends Biochem Sci. 2009. PMID: 19559621 Review.
Cited by
-
Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria.J Biol Chem. 2011 Jun 10;286(23):20292-6. doi: 10.1074/jbc.M110.198523. Epub 2011 Feb 23. J Biol Chem. 2011. PMID: 21345788 Free PMC article.
-
A mechano-chemiosmotic model for the coupling of electron and proton transfer to ATP synthesis in energy-transforming membranes: a personal perspective.Photosynth Res. 2015 Jan;123(1):1-22. doi: 10.1007/s11120-014-0043-3. Epub 2014 Sep 30. Photosynth Res. 2015. PMID: 25266924 Free PMC article.
-
Proteomic response and molecular regulatory mechanisms of Bacillus cereus spores under ultrasound treatment.Ultrason Sonochem. 2021 Oct;78:105732. doi: 10.1016/j.ultsonch.2021.105732. Epub 2021 Aug 21. Ultrason Sonochem. 2021. PMID: 34474268 Free PMC article.
-
The role of ATP in sleep regulation.Front Neurol. 2011 Dec 27;2:87. doi: 10.3389/fneur.2011.00087. eCollection 2011. Front Neurol. 2011. PMID: 22207863 Free PMC article.
-
Dynamic mechanisms driving conformational conversions of the β and ε subunits involved in rotational catalysis of F1-ATPase.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(8):630-647. doi: 10.2183/pjab.93.040. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 29021512 Free PMC article. Review.
References
-
- Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 - PubMed
-
- Pullman M.E., Penefsky H.S., Datta A., Racker E. (1960) Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble, dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 235, 3322–3329 - PubMed
-
- Kagawa Y. (1972) Reconstitution of oxidative phosphorylation. Biochim. Biophys. Acta 265, 297–338 - PubMed
-
- Boyer P.D. (1997) The ATP synthase-a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 - PubMed
-
- Yoshida M., Sone N., Hirata H., Kagawa Y., Ui N. (1979) Subunit structure of adenosine triphosphatase. Comparison of the structure in thermophilic bacterium PS3 with those in mitochondria, chloroplasts, and Escherichia coli. J. Biol. Chem. 254, 9525–9533 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

