Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology
- PMID: 20689598
- PMCID: PMC2912849
- DOI: 10.1371/journal.pone.0011909
Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology
Abstract
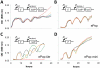
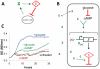
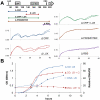
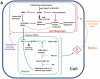
Synthetic biology seeks to enable programmed control of cellular behavior though engineered biological systems. These systems typically consist of synthetic circuits that function inside, and interact with, complex host cells possessing pre-existing metabolic and regulatory networks. Nevertheless, while designing systems, a simple well-defined interface between the synthetic gene circuit and the host is frequently assumed. We describe the generation of robust but unexpected oscillations in the densities of bacterium Escherichia coli populations by simple synthetic suicide circuits containing quorum components and a lysis gene. Contrary to design expectations, oscillations required neither the quorum sensing genes (luxR and luxI) nor known regulatory elements in the P(luxI) promoter. Instead, oscillations were likely due to density-dependent plasmid amplification that established a population-level negative feedback. A mathematical model based on this mechanism captures the key characteristics of oscillations, and model predictions regarding perturbations to plasmid amplification were experimentally validated. Our results underscore the importance of plasmid copy number and potential impact of "hidden interactions" on the behavior of engineered gene circuits - a major challenge for standardizing biological parts. As synthetic biology grows as a discipline, increasing value may be derived from tools that enable the assessment of parts in their final context.
Conflict of interest statement
Figures


Similar articles
-
[Growth and mutation of Escherichia coli with suicide gene circuit based on quorum sensing].Sheng Wu Gong Cheng Xue Bao. 2013 Jun;29(6):726-34. Sheng Wu Gong Cheng Xue Bao. 2013. PMID: 24063233 Chinese.
-
A fast, robust and tunable synthetic gene oscillator.Nature. 2008 Nov 27;456(7221):516-9. doi: 10.1038/nature07389. Epub 2008 Oct 29. Nature. 2008. PMID: 18971928 Free PMC article.
-
Synchronous long-term oscillations in a synthetic gene circuit.Nature. 2016 Oct 27;538(7626):514-517. doi: 10.1038/nature19841. Epub 2016 Oct 12. Nature. 2016. PMID: 27732583 Free PMC article.
-
Decoding biological principles using gene circuits.Mol Biosyst. 2009 Jul;5(7):695-703. doi: 10.1039/b901584c. Epub 2009 May 20. Mol Biosyst. 2009. PMID: 19562108 Review.
-
Quantitative modeling of the interplay between synthetic gene circuits and host physiology: experiments, results, and prospects.Curr Opin Microbiol. 2020 Jun;55:48-56. doi: 10.1016/j.mib.2020.02.008. Epub 2020 Mar 25. Curr Opin Microbiol. 2020. PMID: 32220744 Review.
Cited by
-
A synthetic biology approach to understanding cellular information processing.ACS Synth Biol. 2012 Sep 21;1(9):389-402. doi: 10.1021/sb300044r. ACS Synth Biol. 2012. PMID: 23411668 Free PMC article. Review.
-
Living fabrication of functional semi-interpenetrating polymeric materials.Nat Commun. 2021 Jun 8;12(1):3422. doi: 10.1038/s41467-021-23812-7. Nat Commun. 2021. PMID: 34103521 Free PMC article.
-
Addressing biological uncertainties in engineering gene circuits.Integr Biol (Camb). 2016 Apr 18;8(4):456-64. doi: 10.1039/c5ib00275c. Epub 2015 Dec 17. Integr Biol (Camb). 2016. PMID: 26674800 Free PMC article. Review.
-
Homologous Quorum Sensing Regulatory Circuit: A Dual-Input Genetic Controller for Modulating Quorum Sensing-Mediated Protein Expression in E. coli.ACS Synth Biol. 2020 Oct 16;9(10):2692-2702. doi: 10.1021/acssynbio.0c00179. Epub 2020 Sep 15. ACS Synth Biol. 2020. PMID: 32822530 Free PMC article.
-
Dynamic control and quantification of bacterial population dynamics in droplets.Biomaterials. 2015 Aug;61:239-45. doi: 10.1016/j.biomaterials.2015.05.038. Epub 2015 May 19. Biomaterials. 2015. PMID: 26005763 Free PMC article.
References
-
- Chin JW. Programming and engineering biological networks. Curr Opin Struct Biol. 2006;16:551–556. - PubMed
-
- Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443:527–533. - PubMed
-
- Drubin DA, Way JC, Silver PA. Designing biological systems. Genes Dev. 2007;21:242–254. - PubMed
-
- Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

