Zinc Binding Properties of Engineered RING Finger Domain of Arkadia E3 Ubiquitin Ligase
- PMID: 20689703
- PMCID: PMC2905715
- DOI: 10.1155/2010/323152
Zinc Binding Properties of Engineered RING Finger Domain of Arkadia E3 Ubiquitin Ligase
Abstract
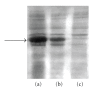
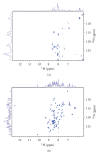
Human Arkadia is a nuclear protein consisted of 989 amino acid residues, with a characteristic RING domain in its C-terminus. The RING domain harbours the E3 ubiquitin ligase activity needed by Arkadia to ubiquitinate its substrates such as negative regulators of TGF-beta signaling. The RING finger domain of Arkadia is a RING-H2 type and its structure and stability is strongly dependent on the presence of two bound Zn(II) ions attached to the protein frame through a defined Cys3-His2-Cys3 motif. In the present paper we transform the RING-H2 type of Arkadia finger domain to nonnative RING sequence, substituting the zinc-binding residues Cys(955) or His(960) to Arginine, through site-directed mutagenesis. The recombinant expression, in Escherichia coli, of the mutants C955R and H960R reveal significant lower yield in respect with the native polypeptide of Arkadia RING-H2 finger domain. In particular, only the C955R mutant exhibits expression yield sufficient for recombinant protein isolation and preliminary studies. Atomic absorption measurements and preliminary NMR data analysis reveal that the C955R point mutation in the RING Finger domain of Arkadia diminishes dramatically the zinc binding affinity, leading to the breakdown of the global structural integrity of the RING construct.
Figures

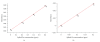
Similar articles
-
High yield expression and NMR characterization of Arkadia E3 ubiquitin ligase RING-H2 finger domain.Biochem Biophys Res Commun. 2009 Jan 16;378(3):498-502. doi: 10.1016/j.bbrc.2008.11.055. Epub 2008 Nov 24. Biochem Biophys Res Commun. 2009. PMID: 19032943
-
A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.J Mol Biol. 2017 Jul 21;429(15):2373-2386. doi: 10.1016/j.jmb.2017.06.012. Epub 2017 Jun 22. J Mol Biol. 2017. PMID: 28647409
-
Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia.Front Mol Biosci. 2022 Feb 23;9:844129. doi: 10.3389/fmolb.2022.844129. eCollection 2022. Front Mol Biosci. 2022. PMID: 35281275 Free PMC article.
-
NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.Proteins. 2012 May;80(5):1484-9. doi: 10.1002/prot.24048. Epub 2012 Mar 13. Proteins. 2012. PMID: 22411132
-
High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides.J Biol Chem. 2003 Apr 25;278(17):15341-8. doi: 10.1074/jbc.M210531200. Epub 2003 Feb 14. J Biol Chem. 2003. PMID: 12588869
Cited by
-
Structural modeling of protein ensembles between E3 RING ligases and SARS-CoV-2: The role of zinc binding domains.J Trace Elem Med Biol. 2023 Jan;75:127089. doi: 10.1016/j.jtemb.2022.127089. Epub 2022 Oct 4. J Trace Elem Med Biol. 2023. PMID: 36209710 Free PMC article.
-
Building Bridges Between Structural and Network-Based Systems Biology.Mol Biotechnol. 2019 Mar;61(3):221-229. doi: 10.1007/s12033-018-0146-8. Mol Biotechnol. 2019. PMID: 30656572 Review.
-
Structural Identification of Metalloproteomes in Marine Diatoms, an Efficient Algae Model in Toxic Metals Bioremediation.Molecules. 2022 Jan 7;27(2):378. doi: 10.3390/molecules27020378. Molecules. 2022. PMID: 35056698 Free PMC article.
-
How to Inactivate Human Ubiquitin E3 Ligases by Mutation.Front Cell Dev Biol. 2020 Feb 4;8:39. doi: 10.3389/fcell.2020.00039. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32117970 Free PMC article. Review.
-
Assessing the Direct Binding of Ark-Like E3 RING Ligases to Ubiquitin and Its Implication on Their Protein Interaction Network.Molecules. 2020 Oct 19;25(20):4787. doi: 10.3390/molecules25204787. Molecules. 2020. PMID: 33086510 Free PMC article.
References
-
- Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Annals of the New York Academy of Sciences. 1993;684:174–192. - PubMed
-
- Freemont PS. RING for destruction? Current Biology. 2000;10(2):R84–R87. - PubMed
-
- Borden KLB, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Current Opinion in Structural Biology. 1996;6(3):395–401. - PubMed
-
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS. Does this have a familiar RING? Trends in Biochemical Sciences. 1996;21(6):208–214. - PubMed
-
- Roehm PC, Berg JM. Sequential metal binding by the RING finger domain of BRCA1. Biochemistry. 1997;36(33):10240–10245. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

