Turning the table: plants consume microbes as a source of nutrients
- PMID: 20689833
- PMCID: PMC2912860
- DOI: 10.1371/journal.pone.0011915
Turning the table: plants consume microbes as a source of nutrients
Abstract
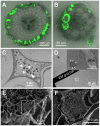
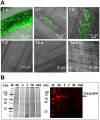
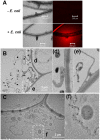
Interactions between plants and microbes in soil, the final frontier of ecology, determine the availability of nutrients to plants and thereby primary production of terrestrial ecosystems. Nutrient cycling in soils is considered a battle between autotrophs and heterotrophs in which the latter usually outcompete the former, although recent studies have questioned the unconditional reign of microbes on nutrient cycles and the plants' dependence on microbes for breakdown of organic matter. Here we present evidence indicative of a more active role of plants in nutrient cycling than currently considered. Using fluorescent-labeled non-pathogenic and non-symbiotic strains of a bacterium and a fungus (Escherichia coli and Saccharomyces cerevisiae, respectively), we demonstrate that microbes enter root cells and are subsequently digested to release nitrogen that is used in shoots. Extensive modifications of root cell walls, as substantiated by cell wall outgrowth and induction of genes encoding cell wall synthesizing, loosening and degrading enzymes, may facilitate the uptake of microbes into root cells. Our study provides further evidence that the autotrophy of plants has a heterotrophic constituent which could explain the presence of root-inhabiting microbes of unknown ecological function. Our discovery has implications for soil ecology and applications including future sustainable agriculture with efficient nutrient cycles.
Conflict of interest statement
Figures



References
-
- Baron C, Zambryski PC. Notes from the underground: Highlights from plant-microbe interactions. Trends Biotechnol. 1995;13:356–362.
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. - PubMed
-
- Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. - PubMed
-
- Triplett EW. Diazotrophic endophytes: Progress and prospects for nitrogen fixation in monocots. Plant Soil. 1996;186:29–38.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

