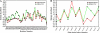T-Analyst: a program for efficient analysis of protein conformational changes by torsion angles
- PMID: 20689979
- PMCID: PMC2940022
- DOI: 10.1007/s10822-010-9376-y
T-Analyst: a program for efficient analysis of protein conformational changes by torsion angles
Abstract
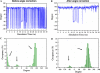
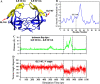
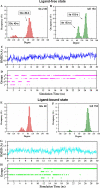
T-Analyst is a user-friendly computer program for analyzing trajectories from molecular modeling. Instead of using Cartesian coordinates for protein conformational analysis, T-Analyst is based on internal bond-angle-torsion coordinates in which internal torsion angle movements, such as side-chain rotations, can be easily detected. The program computes entropy and automatically detects and corrects angle periodicity to produce accurate rotameric states of dihedrals. It also clusters multiple conformations and detects dihedral rotations that contribute hinge-like motions. Correlated motions between selected dihedrals can also be observed from the correlation map. T-Analyst focuses on showing changes in protein flexibility between different states and selecting representative protein conformations for molecular docking studies. The program is provided with instructions and full source code in Perl.
Figures