Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor
- PMID: 20690600
- PMCID: PMC2927733
- DOI: 10.1021/bi1005305
Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor
Abstract
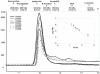
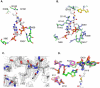
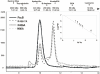
The first committed step of fatty acid and polyketides biosynthesis, the biotin-dependent carboxylation of an acyl-CoA, is catalyzed by acyl-CoA carboxylases (ACCases) such as acetyl-CoA carboxylase (ACC) and propionyl-CoA carboxylase (PCC). ACC and PCC in Streptomyces coelicolor are homologue multisubunit complexes that can carboxylate different short chain acyl-CoAs. While ACC is able to carboxylate acetyl-, propionyl-, or butyryl-CoA with approximately the same specificity, PCC only recognizes propionyl- and butyryl-CoA as substrates. How ACC and PCC have such different specificities toward these substrates is only partially understood. To further understand the molecular basis of how the active site residues can modulate the substrate recognition, we mutated D422, N80, R456, and R457 of PccB, the catalytic beta subunit of PCC. The crystal structures of six PccB mutants and the wild type crystal structure were compared systematically to establish the sequence-structure-function relationship that correlates the observed substrate specificity toward acetyl-, propionyl-, and butyryl-CoA with active site geometry. The experimental data confirmed that D422 is a key determinant of substrate specificity, influencing not only the active site properties but further altering protein stability and causing long-range conformational changes. Mutations of N80, R456, and R457 lead to variations in the quaternary structure of the beta subunit and to a concomitant loss of enzyme activity, indicating the importance of these residues in maintaining the active protein conformation as well as a critical role in substrate binding.
Figures

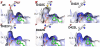
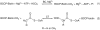
References
-
- Lynen F. New experiments of biotin enzymes. CRC Crit Rev Biochem. 1979;7:103–119. - PubMed
-
- Cronan JE., Jr. The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure id essential and that the domain functions as a dimer. J Biol Chem. 2001;276:37355–37364. - PubMed
-
- Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr. Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. - PubMed
-
- Blanchard CZ, Chapman-Smith A, Wallace JC, Waldrop GL. The biotin domain peptide from the biotin carboxyl carrier protein of Escherichia coli acetyl-CoA carboxylase causes a marked increase in the catalytic efficiency of biotin carboxylase and carboxyltransferase relative to free biotin. J Biol Chem. 1999;274:31767–31769. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

