Regulation of homologous recombination in eukaryotes
- PMID: 20690856
- PMCID: PMC4114321
- DOI: 10.1146/annurev-genet-051710-150955
Regulation of homologous recombination in eukaryotes
Abstract
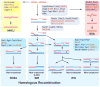
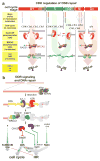
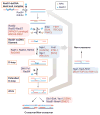
Homologous recombination (HR) is required for accurate chromosome segregation during the first meiotic division and constitutes a key repair and tolerance pathway for complex DNA damage, including DNA double-strand breaks, interstrand crosslinks, and DNA gaps. In addition, recombination and replication are inextricably linked, as recombination recovers stalled and broken replication forks, enabling the evolution of larger genomes/replicons. Defects in recombination lead to genomic instability and elevated cancer predisposition, demonstrating a clear cellular need for recombination. However, recombination can also lead to genome rearrangements. Unrestrained recombination causes undesired endpoints (translocation, deletion, inversion) and the accumulation of toxic recombination intermediates. Evidently, HR must be carefully regulated to match specific cellular needs. Here, we review the factors and mechanistic stages of recombination that are subject to regulation and suggest that recombination achieves flexibility and robustness by proceeding through metastable, reversible intermediates.
Figures

References
-
- Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–67. - PubMed
-
- Alon U. Biological Networks: The tinkerer as an engineer. Science. 2003;301:1866–67. - PubMed
-
- Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

