Chemical-genetic profile analysis of five inhibitory compounds in yeast
- PMID: 20691087
- PMCID: PMC2925817
- DOI: 10.1186/1472-6769-10-6
Chemical-genetic profile analysis of five inhibitory compounds in yeast
Abstract
Background: Chemical-genetic profiling of inhibitory compounds can lead to identification of their modes of action. These profiles can help elucidate the complex interactions between small bioactive compounds and the cell machinery, and explain putative gene function(s).
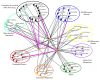
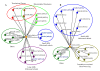
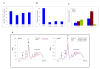
Results: Colony size reduction was used to investigate the chemical-genetic profile of cycloheximide, 3-amino-1,2,4-triazole, paromomycin, streptomycin and neomycin in the yeast Saccharomyces cerevisiae. These compounds target the process of protein biosynthesis. More than 70,000 strains were analyzed from the array of gene deletion mutant yeast strains. As expected, the overall profiles of the tested compounds were similar, with deletions for genes involved in protein biosynthesis being the major category followed by metabolism. This implies that novel genes involved in protein biosynthesis could be identified from these profiles. Further investigations were carried out to assess the activity of three profiled genes in the process of protein biosynthesis using relative fitness of double mutants and other genetic assays.
Conclusion: Chemical-genetic profiles provide insight into the molecular mechanism(s) of the examined compounds by elucidating their potential primary and secondary cellular target sites. Our follow-up investigations into the activity of three profiled genes in the process of protein biosynthesis provided further evidence concerning the usefulness of chemical-genetic analyses for annotating gene functions. We termed these genes TAE2, TAE3 and TAE4 for translation associated elements 2-4.
Figures


References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

