Rate theories for biologists
- PMID: 20691138
- PMCID: PMC3540998
- DOI: 10.1017/S0033583510000120
Rate theories for biologists
Abstract
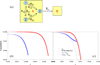
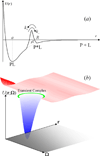
Some of the rate theories that are most useful for modeling biological processes are reviewed. By delving into some of the details and subtleties in the development of the theories, the review will hopefully help the reader gain a more than superficial perspective. Examples are presented to illustrate how rate theories can be used to generate insight at the microscopic level into biomolecular behaviors. An attempt is made to clear up a number of misconceptions in the literature regarding popular rate theories, including the appearance of Planck's constant in the transition-state theory and the Smoluchowski result as an upper limit for protein-protein and protein-DNA association rate constants. Future work in combining the implementation of rate theories through computer simulations with experimental probes of rate processes, and in modeling effects of intracellular environments so that theories can be used for generating rate constants for systems biology studies is particularly exciting.
Figures



Similar articles
-
Studies of proton translocations in biological systems: simulating proton transport in carbonic anhydrase by EVB-based models.Biophys J. 2004 Oct;87(4):2221-39. doi: 10.1529/biophysj.104.043257. Biophys J. 2004. PMID: 15454425 Free PMC article.
-
Surface plasmon resonance: a useful technique for cell biologists to characterize biomolecular interactions.Mol Biol Cell. 2013 Apr;24(7):883-6. doi: 10.1091/mbc.E12-10-0713. Mol Biol Cell. 2013. PMID: 23533209 Free PMC article.
-
Dimensional reduction of Markov state models from renormalization group theory.J Chem Phys. 2016 Sep 28;145(12):124120. doi: 10.1063/1.4963196. J Chem Phys. 2016. PMID: 27782633
-
Modeling for (physical) biologists: an introduction to the rule-based approach.Phys Biol. 2015 Jul 16;12(4):045007. doi: 10.1088/1478-3975/12/4/045007. Phys Biol. 2015. PMID: 26178138 Free PMC article. Review.
-
Theoretical approaches for dynamical ordering of biomolecular systems.Biochim Biophys Acta Gen Subj. 2018 Feb;1862(2):212-228. doi: 10.1016/j.bbagen.2017.10.001. Epub 2017 Oct 6. Biochim Biophys Acta Gen Subj. 2018. PMID: 28988931 Review.
Cited by
-
Contrasting factors on the kinetic path to protein complex formation diminish the effects of crowding agents.Biophys J. 2012 Sep 5;103(5):1011-9. doi: 10.1016/j.bpj.2012.08.009. Biophys J. 2012. PMID: 23009850 Free PMC article.
-
Theory and simulation of diffusion-influenced, stochastically gated ligand binding to buried sites.J Chem Phys. 2011 Oct 14;135(14):145101. doi: 10.1063/1.3645000. J Chem Phys. 2011. PMID: 22010732 Free PMC article.
-
Spectral Rate Theory for Two-State Kinetics.Phys Rev X. 2014 Feb;4(1):011020. doi: 10.1103/PhysRevX.4.011020. Phys Rev X. 2014. PMID: 25356374 Free PMC article.
-
Understanding the Impacts of Conformational Dynamics on the Regulation of Protein-Protein Association by a Multiscale Simulation Method.J Chem Theory Comput. 2020 Aug 11;16(8):5323-5333. doi: 10.1021/acs.jctc.0c00439. Epub 2020 Jul 29. J Chem Theory Comput. 2020. PMID: 32667783 Free PMC article.
-
Classification of protein-protein association rates based on biophysical informatics.BMC Bioinformatics. 2021 Aug 17;22(1):408. doi: 10.1186/s12859-021-04323-0. BMC Bioinformatics. 2021. PMID: 34404340 Free PMC article.
References
-
- Adam G, Delbruck M. Reduction of dimensionality in biological diffusion processes. In: Davidson N, editor. Structural Chemistry and Molecular Biology. San Francisco: W.H. Freeman; 1968. pp. 198–215.
-
- Agmon N. Diffusion with back reaction. Journal of Chemical Physics. 1984;81:2811–2817.
-
- Agmon N, Hopfield JJ. Transient kinetics of chemical reactions with bounded diffusion perpendicular to the reaction coordinate: intramolecular processes with slow conformational changes. Journal of Chemical Physics. 1983;78:6947–6959.
-
- Ai X, Zhou Z, Bai Y, Choy WY. 15N NMR spin relaxation dispersion study of the molecular crowding effects on protein folding under native conditions. Journal of the American Chemical Society. 2006;128:3916–3917. - PubMed
-
- Alm E, Morozov AV, Kortemme T, Baker D. Simple physical models connect theory and experiment in protein folding kinetics. Journal of Molecular Biology. 2002;322:463–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

