PNPASE regulates RNA import into mitochondria
- PMID: 20691904
- PMCID: PMC2921675
- DOI: 10.1016/j.cell.2010.06.035
PNPASE regulates RNA import into mitochondria
Abstract
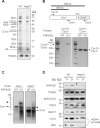
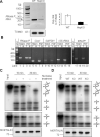
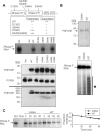
RNA import into mammalian mitochondria is considered essential for replication, transcription, and translation of the mitochondrial genome but the pathway(s) and factors that control this import are poorly understood. Previously, we localized polynucleotide phosphorylase (PNPASE), a 3' --> 5' exoribonuclease and poly-A polymerase, in the mitochondrial intermembrane space, a location lacking resident RNAs. Here, we show a new role for PNPASE in regulating the import of nuclear-encoded RNAs into the mitochondrial matrix. PNPASE reduction impaired mitochondrial RNA processing and polycistronic transcripts accumulated. Augmented import of RNase P, 5S rRNA, and MRP RNAs depended on PNPASE expression and PNPASE-imported RNA interactions were identified. PNPASE RNA processing and import activities were separable and a mitochondrial RNA targeting signal was isolated that enabled RNA import in a PNPASE-dependent manner. Combined, these data strongly support an unanticipated role for PNPASE in mediating the translocation of RNAs into mitochondria.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Mitochondrial matrix reloaded with RNA.Cell. 2010 Aug 6;142(3):362-3. doi: 10.1016/j.cell.2010.07.024. Cell. 2010. PMID: 20691896
References
-
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. - PubMed
-
- Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30:150–155. - PubMed
-
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM061721/GM/NIGMS NIH HHS/United States
- K22CA120147/CA/NCI NIH HHS/United States
- R01 CA107300/CA/NCI NIH HHS/United States
- R01 CA090571/CA/NCI NIH HHS/United States
- R01 GM073981/GM/NIGMS NIH HHS/United States
- R01 CA156674/CA/NCI NIH HHS/United States
- R01CA90571/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- PN2EY018228/EY/NEI NIH HHS/United States
- R01GM073981/GM/NIGMS NIH HHS/United States
- 074454/WT_/Wellcome Trust/United Kingdom
- PN2 EY018228/EY/NEI NIH HHS/United States
- R01 GM061721/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

