A single-nucleotide variation in a p53-binding site affects nutrient-sensitive human SIRT1 expression
- PMID: 20693263
- PMCID: PMC2951863
- DOI: 10.1093/hmg/ddq331
A single-nucleotide variation in a p53-binding site affects nutrient-sensitive human SIRT1 expression
Abstract
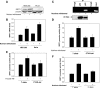
The SIRTUIN1 (SIRT1) deacetylase responds to changes in nutrient availability and regulates mammalian physiology and metabolism. Human and mouse SIRT1 are transcriptionally repressed by p53 via p53 response elements in their proximal promoters. Here, we identify a novel p53-binding sequence in the distal human SIRT1 promoter that is required for nutrient-sensitive SIRT1 transcription. In addition, we show that a common single-nucleotide (C/T) variation in this sequence affects nutrient deprivation-induced SIRT1 transcription, and calorie restriction-induced SIRT1 expression. The p53-binding sequence lies in a region of the SIRT1 promoter that also binds the transcriptional repressor Hypermethylated-In-Cancer-1 (HIC1). Nutrient deprivation increases occupancy by p53, while decreasing occupancy by HIC1, of this region of the promoter. HIC1 and p53 compete with each other for promoter occupancy. In comparison with the T variation, the C variation disrupts the mirror image symmetry of the p53-binding sequence, resulting in decreased binding to p53, decreased nutrient sensitivity of the promoter and impaired calorie restriction-stimulated tissue expression of SIRT1 and SIRT1 target genes AMPKα2 and PGC-1β. Thus, a common SNP in a novel p53-binding sequence in the human SIRT1 promoter affects nutrient-sensitive SIRT1 expression, and could have a significant impact on calorie restriction-induced, SIRT1-mediated, changes in human metabolism and physiology.
Figures

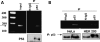


References
-
- Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi:10.1101/gad.13.19.2570. - DOI - PMC - PubMed
-
- Boily G., Seifert E.L., Bevilacqua L., He X.H., Sabourin G., Estey C., Moffat C., Crawford S., Saliba S., Jardine K., et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi:10.1371/journal.pone.0001759. - DOI - PMC - PubMed
-
- Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi:10.1038/nature03354. - DOI - PubMed
-
- Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi:10.1038/nature02583. - DOI - PMC - PubMed
-
- Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.H., Mostoslavsky R., Alt F.W., Wu Z., Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi:10.1038/sj.emboj.7601633. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

