Human SP-A1 (SFTPA1) variant-specific 3' UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene
- PMID: 20693318
- PMCID: PMC2957414
- DOI: 10.1152/ajplung.00113.2010
Human SP-A1 (SFTPA1) variant-specific 3' UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene
Abstract
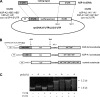
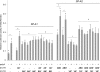
Human surfactant protein A (SP-A) is encoded by two functional genes (SFTPA1, SFTPA2) with a high degree of sequence identity. Sequence differences among these genes and their genetic variants have been observed at the 5' and 3' untranslated regions (UTRs). In this work, we studied the impact on translation of the SFTPA1 (hSP-A1) and SFTPA2 (hSP-A2) gene 5' UTR splice variants and 3' UTR sequence variants, in the presence or absence of poly(A) tail. We generated constructs containing the luciferase reporter gene flanked upstream by one of the hSP-A 5' UTR splice variants and/or downstream by one hSP-A 3' UTR sequence variant. mRNA transcripts were prepared by in vitro transcription and used for either in vitro translation with a rabbit reticulocyte lysate or transient transfection of the lung adenocarcinoma cell line NCI-H441. The luciferase activity results indicate that hSP-A 5' UTR and 3' UTR together have an additive effect on translation. In this context, the hSP-A1 6A(3) and 6A(4) 3' UTR variants exhibited higher translation efficiency than the 6A(2) variant (P <0.05), whereas no significant difference was observed between the two hSP-A2 3' UTRs studied (1A(0), 1A(3)). Further sequence analysis revealed that a deletion of an 11-nucleotide (nt) element in both the 6A(3) and 6A(4) 3' UTR variants changes the predicted secondary structure stability and the number of putative miRNA binding sites. Removal of this 11-nt element in the 6A(2) 3' UTR resulted in increased translation, and the opposite effect was observed when the 11-nt element was cloned in a guest 3' UTR (6A(3), 6A(4)). These results indicate that sequence differences among hSP-A gene variants may account for differential regulation at the translational level.
Figures






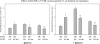
References
-
- Alexiou P, Maragkakis M, Papadopoulos G, Reczko M, Hatzigeorgiou A. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 25: 3049–3055, 2009 - PubMed
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474, 2009 - PubMed
-
- Boggaram V, Mendelson C. Transcriptional regulation of the gene encoding the major surfactant protein (SP-A) in rabbit fetal lung. J Biol Chem 263: 19060–19065, 1988 - PubMed
-
- Boggaram V, Smith M, Mendelson C. Posttranscriptional regulation of surfactant protein-A messenger RNA in human fetal lung in vitro by glucocorticoids. Mol Endocrinol 5: 414–423, 1991 - PubMed
-
- Bruns G, Stroh H, Veldman G, Latt S, Floros J. The 35 kd pulmonary surfactant-associated protein is encoded on chromosome 10. Hum Genet 76: 58–62, 1987 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

