N-glycosylation augmentation of the cystic fibrosis epithelium improves Pseudomonas aeruginosa clearance
- PMID: 20693405
- PMCID: PMC3135844
- DOI: 10.1165/rcmb.2009-0285OC
N-glycosylation augmentation of the cystic fibrosis epithelium improves Pseudomonas aeruginosa clearance
Abstract
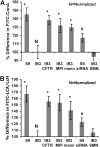
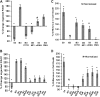
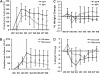
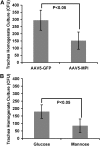
Chronic lung colonization with Pseudomonas aeruginosa is anticipated in cystic fibrosis (CF). Abnormal terminal glycosylation has been implicated as a candidate for this condition. We previously reported a down-regulation of mannose-6-phosphate isomerase (MPI) for core N-glycan production in the CFTR-defective human cell line (IB3). We found a 40% decrease in N-glycosylation of IB3 cells compared with CFTR-corrected human cell line (S9), along with a threefold-lower surface attachment of P. aeruginosa strain, PAO1. There was a twofold increase in intracellular bacteria in S9 cells compared with IB3 cells. After a 4-hour clearance period, intracellular bacteria in IB3 cells increased twofold. Comparatively, a twofold decrease in intracellular bacteria occurred in S9 cells. Gene augmentation in IB3 cells with hMPI or hCFTR reversed these IB3 deficiencies. Mannose-6-phosphate can be produced from external mannose independent of MPI, and correction in the IB3 clearance deficiencies was observed when cultured in mannose-rich medium. An in vivo model for P. aeruginosa colonization in the upper airways revealed an increased bacterial burden in the trachea and oropharynx of nontherapeutic CF mice compared with mice treated either with an intratracheal delivery adeno-associated viral vector 5 expressing murine MPI, or a hypermannose water diet. Finally, a modest lung inflammatory response was observed in CF mice, and was partially corrected by both treatments. Augmenting N-glycosylation to attenuate colonization of P. aeruginosa in CF airways reveals a new therapeutic avenue for a hallmark disease condition in CF.
Figures


Similar articles
-
Defective acid sphingomyelinase pathway with Pseudomonas aeruginosa infection in cystic fibrosis.Am J Respir Cell Mol Biol. 2009 Sep;41(3):367-75. doi: 10.1165/rcmb.2008-0295OC. Epub 2009 Jan 23. Am J Respir Cell Mol Biol. 2009. PMID: 19168701 Free PMC article.
-
MPB-07 reduces the inflammatory response to Pseudomonas aeruginosa in cystic fibrosis bronchial cells.Am J Respir Cell Mol Biol. 2007 May;36(5):615-24. doi: 10.1165/rcmb.2006-0200OC. Epub 2006 Dec 29. Am J Respir Cell Mol Biol. 2007. PMID: 17197571
-
Dendrimer-based selective autophagy-induction rescues ΔF508-CFTR and inhibits Pseudomonas aeruginosa infection in cystic fibrosis.PLoS One. 2017 Sep 13;12(9):e0184793. doi: 10.1371/journal.pone.0184793. eCollection 2017. PLoS One. 2017. PMID: 28902888 Free PMC article.
-
The role of the CFTR in susceptibility to Pseudomonas aeruginosa infections in cystic fibrosis.Trends Microbiol. 2000 Nov;8(11):514-20. doi: 10.1016/s0966-842x(00)01872-2. Trends Microbiol. 2000. PMID: 11121762 Review.
-
Harnessing Neutrophil Survival Mechanisms during Chronic Infection by Pseudomonas aeruginosa: Novel Therapeutic Targets to Dampen Inflammation in Cystic Fibrosis.Front Cell Infect Microbiol. 2017 Jun 30;7:243. doi: 10.3389/fcimb.2017.00243. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28713772 Free PMC article. Review.
Cited by
-
Mimicking the host and its microenvironment in vitro for studying mucosal infections by Pseudomonas aeruginosa.Pathog Dis. 2014 Jun;71(1):1-19. doi: 10.1111/2049-632X.12180. Epub 2014 May 23. Pathog Dis. 2014. PMID: 24737619 Free PMC article. Review.
-
Development of rAAV2-CFTR: History of the First rAAV Vector Product to be Used in Humans.Hum Gene Ther Methods. 2016 Apr;27(2):49-58. doi: 10.1089/hgtb.2015.150. Epub 2016 Feb 19. Hum Gene Ther Methods. 2016. PMID: 26895204 Free PMC article. Review.
-
Infection, inflammation and host carbohydrates: a Glyco-Evasion Hypothesis.Glycobiology. 2012 Aug;22(8):1019-30. doi: 10.1093/glycob/cws070. Epub 2012 Apr 5. Glycobiology. 2012. PMID: 22492234 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

