The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture
- PMID: 20693407
- PMCID: PMC3101955
- DOI: 10.1074/mcp.M110.002915
The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture
Abstract
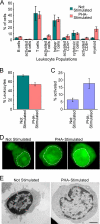
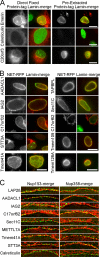
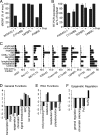
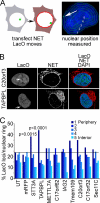
A favored hypothesis to explain the pathology underlying nuclear envelopathies is that mutations in nuclear envelope proteins alter genome/chromatin organization and thus gene expression. To identify nuclear envelope proteins that play roles in genome organization, we analyzed nuclear envelopes from resting and phytohemagglutinin-activated leukocytes because leukocytes have a particularly high density of peripheral chromatin that undergoes significant reorganization upon such activation. Thus, nuclear envelopes were isolated from leukocytes in the two states and analyzed by multidimensional protein identification technology using an approach that used expected contaminating membranes as subtractive fractions. A total of 3351 proteins were identified between both nuclear envelope data sets among which were 87 putative nuclear envelope transmembrane proteins (NETs) that were not identified in a previous proteomics analysis of liver nuclear envelopes. Nuclear envelope localization was confirmed for 11 new NETs using tagged fusion proteins and antibodies on spleen cryosections. 27% of the new proteins identified were unique to one or the other of the two leukocyte states. Differences in expression between activated and resting leukocytes were confirmed for some NETs by RT-PCR, and most of these proteins appear to only be expressed in certain types of blood cells. Several known proteins identified in both data sets have functions in chromatin organization and gene regulation. To test whether the novel NETs identified might include those that also regulate chromatin, nine were run through two screens for different chromatin effects. One screen found two NETs that can recruit a specific gene locus to the nuclear periphery, and the second found a different NET that promotes chromatin condensation. The variation in the protein milieu with pharmacological activation of the same cell population and consequences for gene regulation suggest that the nuclear envelope is a complex regulatory system with significant influences on genome organization.
Figures




Similar articles
-
The nuclear envelope proteome differs notably between tissues.Nucleus. 2012 Nov-Dec;3(6):552-64. doi: 10.4161/nucl.22257. Epub 2012 Sep 18. Nucleus. 2012. PMID: 22990521 Free PMC article.
-
Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis.BMC Cell Biol. 2006 Oct 24;7:38. doi: 10.1186/1471-2121-7-38. BMC Cell Biol. 2006. PMID: 17062158 Free PMC article.
-
Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations.Mol Cell Proteomics. 2011 Jan;10(1):M110.003129. doi: 10.1074/mcp.M110.003129. Epub 2010 Sep 27. Mol Cell Proteomics. 2011. PMID: 20876400 Free PMC article.
-
NET gains and losses: the role of changing nuclear envelope proteomes in genome regulation.Curr Opin Cell Biol. 2014 Jun;28:105-20. doi: 10.1016/j.ceb.2014.04.005. Epub 2014 Jun 2. Curr Opin Cell Biol. 2014. PMID: 24886773 Review.
-
Identification of novel integral membrane proteins of the nuclear envelope with potential disease links using subtractive proteomics.Novartis Found Symp. 2005;264:63-76; discussion 76-80, 227-30. Novartis Found Symp. 2005. PMID: 15773748 Review.
Cited by
-
Nuclear envelope and genome interactions in cell fate.Front Genet. 2015 Mar 19;6:95. doi: 10.3389/fgene.2015.00095. eCollection 2015. Front Genet. 2015. PMID: 25852741 Free PMC article. Review.
-
The nuclear envelope proteome differs notably between tissues.Nucleus. 2012 Nov-Dec;3(6):552-64. doi: 10.4161/nucl.22257. Epub 2012 Sep 18. Nucleus. 2012. PMID: 22990521 Free PMC article.
-
Emerin in health and disease.Semin Cell Dev Biol. 2014 May;29:95-106. doi: 10.1016/j.semcdb.2013.12.008. Epub 2013 Dec 21. Semin Cell Dev Biol. 2014. PMID: 24365856 Free PMC article. Review.
-
Identification of new transmembrane proteins concentrated at the nuclear envelope using organellar proteomics of mesenchymal cells.Nucleus. 2019 Dec;10(1):126-143. doi: 10.1080/19491034.2019.1618175. Nucleus. 2019. PMID: 31142202 Free PMC article.
-
Native lamin A/C proteomes and novel partners from heart and skeletal muscle in a mouse chronic inflammation model of human frailty.Front Cell Dev Biol. 2023 Oct 23;11:1240285. doi: 10.3389/fcell.2023.1240285. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37936983 Free PMC article.
References
-
- Schirmer E. C., Foisner R. (2007) Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 313, 2167–2179 - PubMed
-
- Suntharalingam M., Wente S. R. (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775–789 - PubMed
-
- Callan H. G., Tomlin S. G. (1950) Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc. R. Soc. Lond. B Biol. Sci. 137, 367–378 - PubMed
-
- Starr D. A., Fischer J. A. (2005) KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136–1146 - PubMed
-
- Stewart C. L., Roux K. J., Burke B. (2007) Blurring the boundary: the nuclear envelope extends its reach. Science 318, 1408–1412 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

