Mutation of high-affinity methionine permease contributes to selenomethionyl protein production in Saccharomyces cerevisiae
- PMID: 20693451
- PMCID: PMC2950440
- DOI: 10.1128/AEM.01026-10
Mutation of high-affinity methionine permease contributes to selenomethionyl protein production in Saccharomyces cerevisiae
Abstract
The production of selenomethionine (SeMet) derivatives of recombinant proteins allows phase determination by single-wavelength or multiwavelength anomalous dispersion phasing in X-ray crystallography, and this popular approach has permitted the crystal structures of numerous proteins to be determined. Although yeast is an ideal host for the production of large amounts of eukaryotic proteins that require posttranslational modification, the toxic effects of SeMet often interfere with the preparation of protein derivatives containing this compound. We previously isolated a mutant strain (SMR-94) of the methylotrophic yeast Pichia pastoris that is resistant to both SeMet and selenate and demonstrated its applicability for the production of proteins suitable for X-ray crystallographic analysis. However, the molecular basis for resistance to SeMet by the SMR-94 strain remains unclear. Here, we report the characterization of SeMet-resistant mutants of Saccharomyces cerevisiae and the identification of a mutant allele of the MUP1 gene encoding high-affinity methionine permease, which confers SeMet resistance. Although the total methionine uptake by the mup1 mutant (the SRY5-7 strain) decreased to 47% of the wild-type level, it was able to incorporate SeMet into the overexpressed epidermal growth factor peptide with 73% occupancy, indicating the importance of the moderate uptake of SeMet by amino acid permeases other than Mup1p for the alleviation of SeMet toxicity. In addition, under standard culture conditions, the mup1 mutant showed higher productivity of the SeMet derivative relative to other SeMet-resistant mutants. Based on these results, we conclude that the mup1 mutant would be useful for the preparation of selenomethionyl proteins for X-ray crystallography.
Figures
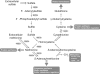

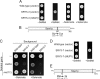

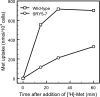


References
-
- Budisa, N., R. Huber, R. Golbik, C. Minks, E. Weyher, and L. Moroder. 1998. Atomic mutations in annexin V—thermodynamic studies of isomorphous protein variants. Eur. J. Biochem. 253:1-9. - PubMed
-
- Bushnell, D. A., P. Cramer, and R. D. Kornberg. 2001. Selenomethionine incorporation in Saccharomyces cerevisiae RNA polymerase II. Structure 9:R11-R14. - PubMed
-
- Cherest, H., and Y. Surdin-Kerjan. 1978. S-Adenosyl methionine requiring mutants in Saccharomyces cerevisiae: evidences for the existence of two methionine adenosyl transferases. Mol. Gen. Genet. 163:153-167. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

