CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors
- PMID: 20693478
- PMCID: PMC2963815
- DOI: 10.1101/gr.109405.110
CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors
Abstract
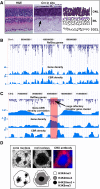
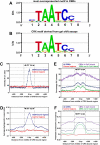
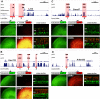
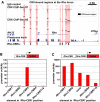
Approximately 98% of mammalian DNA is noncoding, yet we understand relatively little about the function of this enigmatic portion of the genome. The cis-regulatory elements that control gene expression reside in noncoding regions and can be identified by mapping the binding sites of tissue-specific transcription factors. Cone-rod homeobox (CRX) is a key transcription factor in photoreceptor differentiation and survival, but its in vivo targets are largely unknown. Here, we used chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) on CRX to identify thousands of cis-regulatory regions around photoreceptor genes in adult mouse retina. CRX directly regulates downstream photoreceptor transcription factors and their target genes via a network of spatially distributed regulatory elements around each locus. CRX-bound regions act in a synergistic fashion to activate transcription and contain multiple CRX binding sites which interact in a spacing- and orientation-dependent manner to fine-tune transcript levels. CRX ChIP-seq was also performed on Nrl(-/-) retinas, which represent an enriched source of cone photoreceptors. Comparison with the wild-type ChIP-seq data set identified numerous rod- and cone-specific CRX-bound regions as well as many shared elements. Thus, CRX combinatorially orchestrates the transcriptional networks of both rods and cones by coordinating the expression of photoreceptor genes including most retinal disease genes. In addition, this study pinpoints thousands of noncoding regions of relevance to both Mendelian and complex retinal disease.
Figures

References
-
- Bateman E 1998. Autoregulation of eukaryotic transcription factors. Prog Nucleic Acid Res Mol Biol 60: 133–168 - PubMed
-
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS 1999. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet 21: 355–356 - PubMed
-
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL 2001. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107: 579–589 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
