The type II restriction endonuclease MvaI has dual specificity
- PMID: 20693529
- PMCID: PMC3001055
- DOI: 10.1093/nar/gkq676
The type II restriction endonuclease MvaI has dual specificity
Abstract
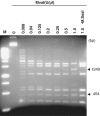
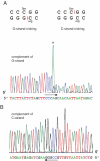
The MvaI restriction endonuclease cuts 5'-CC↓AGG-3'/5'-CC↑TGG-3' sites as indicated by the arrows. N4-methylation of the inner cytosines (C(m4)CAGG/C(m4)CTGG) protects the site against MvaI cleavage. Here, we show that MvaI nicks the G-strand of the related sequence (CCGGG/CCCGG, BcnI site) if the inner cytosines are C5-methylated: C(m5)C↓GGG/CC(m5)CGG. At M.SssI-methylated SmaI sites, where two oppositely oriented methylated BcnI sites partially overlap, double-nicking leads to double-strand cleavage (CC(m5)C↓GGG/CC(m5)C↑GGG) generating fragments with blunt ends. The double-strand cleavage rate and the stringency of substrate site recognition is lower at the methylation-dependent site than at the canonical target site. MvaI is the first restriction endonuclease shown to possess, besides the 'normal' activity on its unmethylated recognition site, also a methylation-directed activity on a different sequence.
Figures



Similar articles
-
Degenerate sequence recognition by the monomeric restriction enzyme: single mutation converts BcnI into a strand-specific nicking endonuclease.Nucleic Acids Res. 2011 May;39(9):3744-53. doi: 10.1093/nar/gkq1351. Epub 2011 Jan 11. Nucleic Acids Res. 2011. PMID: 21227928 Free PMC article.
-
Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.Nucleic Acids Res. 2007;35(6):2035-46. doi: 10.1093/nar/gkm064. Epub 2007 Mar 7. Nucleic Acids Res. 2007. PMID: 17344322 Free PMC article.
-
[Cleavage by restriction endonucleases MvaI and EcoRII of substrates modified in amino groups of heterocyclic bases].Bioorg Khim. 1990 Apr;16(4):501-6. Bioorg Khim. 1990. PMID: 2375778 Russian.
-
Substrate recognition by the Pvu II endonuclease: binding and cleavage of CAG5mCTG sites.Nucleic Acids Res. 1999 Feb 15;27(4):1032-8. doi: 10.1093/nar/27.4.1032. Nucleic Acids Res. 1999. PMID: 9927736 Free PMC article.
-
Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity.Nucleic Acids Res. 2011 Jan;39(1):1-18. doi: 10.1093/nar/gkq742. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805246 Free PMC article. Review.
Cited by
-
Cytosine-to-uracil deamination by SssI DNA methyltransferase.PLoS One. 2013 Oct 21;8(10):e79003. doi: 10.1371/journal.pone.0079003. eCollection 2013. PLoS One. 2013. PMID: 24205358 Free PMC article.
-
Cloning and characterization of the TneDI restriction: modification system of Thermotoga neapolitana.Extremophiles. 2011 Nov;15(6):665-72. doi: 10.1007/s00792-011-0397-9. Epub 2011 Sep 15. Extremophiles. 2011. PMID: 21918796
References
-
- Smith HO, Wilcox KW. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J. Mol. Biol. 1970;51:379–391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

