Residual tumor cells are unique cellular targets in glioblastoma
- PMID: 20695020
- PMCID: PMC4445859
- DOI: 10.1002/ana.22036
Residual tumor cells are unique cellular targets in glioblastoma
Abstract
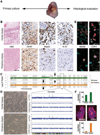
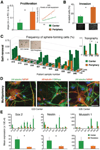
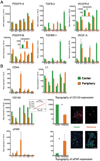
Residual tumor cells remain beyond the margins of every glioblastoma (GBM) resection. Their resistance to postsurgical therapy is considered a major driving force of mortality, but their biology remains largely uncharacterized. In this study, residual tumor cells were derived via experimental biopsy of the resection margin after standard neurosurgery for direct comparison with samples from the routinely resected tumor tissue. In vitro analysis of proliferation, invasion, stem cell qualities, GBM-typical antigens, genotypes, and in vitro drug and irradiation challenge studies revealed these cells as unique entities. Our findings suggest a need for characterization of residual tumor cells to optimize diagnosis and treatment of GBM.
Conflict of interest statement
Nothing to report.
Figures
References
-
- Mitchell P, Ellison DW, Mendelow AD. Surgery for malignant gliomas: mechanistic reasoning and slippery statistics. Lancet Neurol. 2005;4:413–422. - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. - PubMed
-
- Stummer W, Pichlmeier U, Meinel T, et al. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7:392–401. - PubMed
-
- Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. - PubMed
-
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

