eGIFT: mining gene information from the literature
- PMID: 20696046
- PMCID: PMC2929241
- DOI: 10.1186/1471-2105-11-418
eGIFT: mining gene information from the literature
Abstract
Background: With the biomedical literature continually expanding, searching PubMed for information about specific genes becomes increasingly difficult. Not only can thousands of results be returned, but gene name ambiguity leads to many irrelevant hits. As a result, it is difficult for life scientists and gene curators to rapidly get an overall picture about a specific gene from documents that mention its names and synonyms.
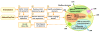
Results: In this paper, we present eGIFT (http://biotm.cis.udel.edu/eGIFT), a web-based tool that associates informative terms, called iTerms, and sentences containing them, with genes. To associate iTerms with a gene, eGIFT ranks iTerms about the gene, based on a score which compares the frequency of occurrence of a term in the gene's literature to its frequency of occurrence in documents about genes in general. To retrieve a gene's documents (Medline abstracts), eGIFT considers all gene names, aliases, and synonyms. Since many of the gene names can be ambiguous, eGIFT applies a disambiguation step to remove matches that do not correspond to this gene. Another additional filtering process is applied to retain those abstracts that focus on the gene rather than mention it in passing. eGIFT's information for a gene is pre-computed and users of eGIFT can search for genes by using a name or an EntrezGene identifier. iTerms are grouped into different categories to facilitate a quick inspection. eGIFT also links an iTerm to sentences mentioning the term to allow users to see the relation between the iTerm and the gene. We evaluated the precision and recall of eGIFT's iTerms for 40 genes; between 88% and 94% of the iTerms were marked as salient by our evaluators, and 94% of the UniProtKB keywords for these genes were also identified by eGIFT as iTerms.
Conclusions: Our evaluations suggest that iTerms capture highly-relevant aspects of genes. Furthermore, by showing sentences containing these terms, eGIFT can provide a quick description of a specific gene. eGIFT helps not only life scientists survey results of high-throughput experiments, but also annotators to find articles describing gene aspects and functions.
Figures
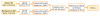

References
-
- McEntyre J, Lipman D. PubMed: bridging the information gap. Canadian Medical Association Journal. 2001;164(9):1317–1319. http://www.ncbi.nlm.nih.gov/sites/entrez - PMC - PubMed
-
- BioMed Central. http://www.biomedcentral.com/
-
- Liu Y, Brandon M, Navathe S, Dingledine R, Ciliax BJ. Text mining functional keywords associated with genes. MedInfo. 2004;11:292–296. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

