Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci
- PMID: 20696051
- PMCID: PMC2928199
- DOI: 10.1186/1471-2180-10-213
Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci
Abstract
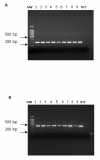
Background: Cryptosporidium is a protozoan parasite that causes diarrheal illness in a wide range of hosts including humans. Two species, C. parvum and C. hominis are of primary public health relevance. Genome sequences of these two species are available and show only 3-5% sequence divergence. We investigated this sequence variability, which could correspond either to sequence gaps in the published genome sequences or to the presence of species-specific genes. Comparative genomic tools were used to identify putative species-specific genes and a subset of these genes was tested by PCR in a collection of Cryptosporidium clinical isolates and reference strains.
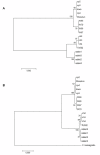
Results: The majority of the putative species-specific genes examined were in fact common to C. parvum and C. hominis. PCR product sequence analysis revealed interesting SNPs, the majority of which were species-specific. These genetic loci allowed us to construct a robust and multi-locus analysis. The Neighbour-Joining phylogenetic tree constructed clearly discriminated the previously described lineages of Cryptosporidium species and subtypes.
Conclusions: Most of the genes identified as being species specific during bioinformatics in Cryptosporidium sp. are in fact present in multiple species and only appear species specific because of gaps in published genome sequences. Nevertheless SNPs may offer a promising approach to studying the taxonomy of closely related species of Cryptosporidia.
Figures
Similar articles
-
Increased diversity and novel subtypes among clinical Cryptosporidium parvum and Cryptosporidium hominis isolates in Southern Ireland.Exp Parasitol. 2020 Nov;218:107967. doi: 10.1016/j.exppara.2020.107967. Epub 2020 Aug 25. Exp Parasitol. 2020. PMID: 32858044
-
PCR slippage across the ML-2 microsatellite of the Cryptosporidium MIC1 locus enables development of a PCR assay capable of distinguishing the zoonotic Cryptosporidium parvum from other human infectious Cryptosporidium species.Zoonoses Public Health. 2014 Aug;61(5):324-37. doi: 10.1111/zph.12074. Epub 2013 Aug 17. Zoonoses Public Health. 2014. PMID: 23954136
-
Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species.BMC Genomics. 2019 May 22;20(1):406. doi: 10.1186/s12864-019-5788-9. BMC Genomics. 2019. PMID: 31117941 Free PMC article.
-
Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of "next generation" technologies--research review.Biotechnol Adv. 2010 Jan-Feb;28(1):17-26. doi: 10.1016/j.biotechadv.2009.08.003. Biotechnol Adv. 2010. PMID: 19699288 Review.
-
Cryptosporidium hominis infections in non-human animal species: revisiting the concept of host specificity.Int J Parasitol. 2020 Apr;50(4):253-262. doi: 10.1016/j.ijpara.2020.01.005. Epub 2020 Mar 20. Int J Parasitol. 2020. PMID: 32205089 Review.
Cited by
-
Giardia/Cryptosporidium QUIK CHEK Assay Is More Specific Than Quantitative Polymerase Chain Reaction for Rapid Point-of-care Diagnosis of Cryptosporidiosis in Infants in Bangladesh.Clin Infect Dis. 2018 Nov 28;67(12):1897-1903. doi: 10.1093/cid/ciy372. Clin Infect Dis. 2018. PMID: 29718129 Free PMC article.
-
Generation of whole genome sequences of new Cryptosporidium hominis and Cryptosporidium parvum isolates directly from stool samples.BMC Genomics. 2015 Aug 29;16(1):650. doi: 10.1186/s12864-015-1805-9. BMC Genomics. 2015. PMID: 26318339 Free PMC article.
-
First detection of Cryptosporidium DNA in blood and cerebrospinal fluid of HIV-infected patients.Parasitol Res. 2018 Mar;117(3):875-881. doi: 10.1007/s00436-018-5766-1. Epub 2018 Feb 6. Parasitol Res. 2018. PMID: 29411108
-
Subtyping of Cryptosporidium cuniculus and genotyping of Enterocytozoon bieneusi in rabbits in two farms in Heilongjiang Province, China.Parasite. 2016;23:52. doi: 10.1051/parasite/2016063. Epub 2016 Nov 24. Parasite. 2016. PMID: 27882867 Free PMC article.
-
Under-notification of cryptosporidiosis by routine clinical and laboratory practices among non-hospitalised children with acute diarrhoea in Southern Spain.Infection. 2012 Apr;40(2):113-9. doi: 10.1007/s15010-011-0188-3. Epub 2011 Sep 6. Infection. 2012. PMID: 21898121
References
-
- Cacciò S. Molecular epidemiology of human cryptosporidiosis. Parassitologia. 2005;47:185–192. - PubMed
-
- Morgan UM, Deplazes P, Forbes DA, Spano F, Hertzberg H, Sargent KD, Elliot A, Thompson RC. Sequence and PCR-RFLP analysis of the internal transcribed spacers of the rDNA repeat unit in isolates of Cryptosporidium from different hosts. Parasitology. 1999;118(Pt 1):49–58. doi: 10.1017/S0031182098003412. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

