Untangling spider silk evolution with spidroin terminal domains
- PMID: 20696068
- PMCID: PMC2928236
- DOI: 10.1186/1471-2148-10-243
Untangling spider silk evolution with spidroin terminal domains
Abstract
Background: Spidroins are a unique family of large, structural proteins that make up the bulk of spider silk fibers. Due to the highly variable nature of their repetitive sequences, spidroin evolutionary relationships have principally been determined from their non-repetitive carboxy (C)-terminal domains, though they offer limited character data. The few known spidroin amino (N)-terminal domains have been difficult to obtain, but potentially contain critical phylogenetic information for reconstructing the diversification of spider silks. Here we used silk gland expression data (ESTs) from highly divergent species to evaluate the functional significance and phylogenetic utility of spidroin N-terminal domains.
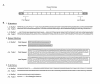
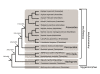
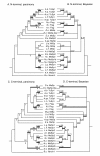
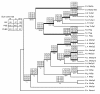
Results: We report 11 additional spidroin N-termini found by sequencing approximately 1,900 silk gland cDNAs from nine spider species that shared a common ancestor > 240 million years ago. In contrast to their hyper-variable repetitive regions, spidroin N-terminal domains have retained striking similarities in sequence identity, predicted secondary structure, and hydrophobicity. Through separate and combined phylogenetic analyses of N-terminal domains and their corresponding C-termini, we find that combined analysis produces the most resolved trees and that N-termini contribute more support and less conflict than the C-termini. These analyses show that paralogs largely group by silk gland type, except for the major ampullate spidroins. Moreover, spidroin structural motifs associated with superior tensile strength arose early in the history of this gene family, whereas a motif conferring greater extensibility convergently evolved in two distantly related paralogs.
Conclusions: A non-repetitive N-terminal domain appears to be a universal attribute of spidroin proteins, likely retained from the origin of spider silk production. Since this time, spidroin N-termini have maintained several features, consistent with this domain playing a key role in silk assembly. Phylogenetic analyses of the conserved N- and C-terminal domains illustrate dramatic radiation of the spidroin gene family, involving extensive duplications, shifts in expression patterns and extreme diversification of repetitive structural sequences that endow spider silks with an unparalleled range of mechanical properties.
Figures
References
-
- Swanson BO, Blackledge TA, Summers AP, Hayashi CY. Spider dragline silk: correlated and mosaic evolution in high-performance biological materials. Evolution. 2006;60:2539–2551. - PubMed
-
- Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: from fibroin sequence to mechanical function. J Exp Biol. 1999;202:3295–3303. - PubMed
-
- Hinman MB, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J Biol Chem. 1992;267:19320–19324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

