Nuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells
- PMID: 20696077
- PMCID: PMC3087322
- DOI: 10.1186/1471-2407-10-414
Nuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells
Abstract
Background: In breast cancer cells, the metastatic cell state is strongly correlated to epithelial-to-mesenchymal transition (EMT) and the CD44+/CD24- stem cell phenotype. However, the MCF-7 cell line, which has a luminal epithelial-like phenotype and lacks a CD44+/CD24- subpopulation, has rare cell populations with higher Matrigel invasive ability. Thus, what are the potentially important differences between invasive and non-invasive breast cancer cells, and are the differences related to EMT or CD44/CD24 expression?
Methods: Throughout the sequential selection process using Matrigel, we obtained MCF-7-14 cells of opposite migratory and invasive capabilities from MCF-7 cells. Comparative analysis of epithelial and mesenchymal marker expression was performed between parental MCF-7, selected MCF-7-14, and aggressive mesenchymal MDA-MB-231 cells. Furthermore, using microarray expression profiles of these cells, we selected differentially expressed genes for their invasive potential, and performed pathway and network analysis to identify a set of interesting genes, which were evaluated by RT-PCR, flow cytometry or function-blocking antibody treatment.
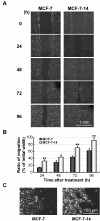
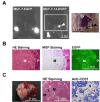
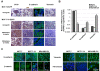
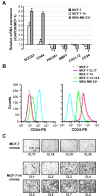
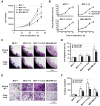
Results: MCF-7-14 cells had enhanced migratory and invasive ability compared with MCF-7 cells. Although MCF-7-14 cells, similar to MCF-7 cells, expressed E-cadherin but neither vimentin nor fibronectin, beta-catenin was expressed not only on the cell membrane but also in the nucleus. Furthermore, using gene expression profiles of MCF-7, MCF-7-14 and MDA-MB-231 cells, we demonstrated that MCF-7-14 cells have alterations in signaling pathways regulating cell migration and identified a set of genes (PIK3R1, SOCS2, BMP7, CD44 and CD24). Interestingly, MCF-7-14 and its invasive clone CL6 cells displayed increased CD44 expression and downregulated CD24 expression compared with MCF-7 cells. Anti-CD44 antibody treatment significantly decreased cell migration and invasion in both MCF-7-14 and MCF-7-14 CL6 cells as well as MDA-MB-231 cells.
Conclusions: MCF-7-14 cells are a novel model for breast cancer metastasis without requiring constitutive EMT and are categorized as a "metastable phenotype", which can be distinguished from both epithelial and mesenchymal cells. The alterations and characteristics of MCF-7-14 cells, especially nuclear beta-catenin and CD44 upregulation, may characterize invasive cell populations in breast cancer.
Figures

References
-
- Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix MJ, Wu R, Wu CW. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17:353–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

