Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems
- PMID: 20696400
- PMCID: PMC8229532
- DOI: 10.1016/j.str.2010.04.018
Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems
Abstract
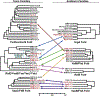
Bacterial toxin-antitoxin (TA) systems serve a variety of physiological functions including regulation of cell growth and maintenance of foreign genetic elements. Sequence analyses suggest that TA families are linked by complex evolutionary relationships reflecting likely swapping of functional domains between different TA families. Our crystal structures of Phd-Doc from bacteriophage P1, the HigA antitoxin from Escherichia coli CFT073, and YeeU of the YeeUWV systems from E. coli K12 and Shigella flexneri confirm this inference and reveal additional, unanticipated structural relationships. The growth-regulating Doc toxin exhibits structural similarity to secreted virulence factors that are toxic for eukaryotic target cells. The Phd antitoxin possesses the same fold as both the YefM and NE2111 antitoxins that inhibit structurally unrelated toxins. YeeU, which has an antitoxin-like activity that represses toxin expression, is structurally similar to the ribosome-interacting toxins YoeB and RelE. These observations suggest extensive functional exchanges have occurred between TA systems during bacterial evolution.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
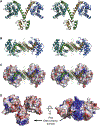
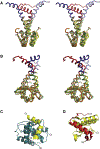
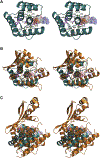
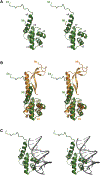

References
-
- Ades SE, and Sauer RT (1995). Specificity of minor-groove and majorgroove interactions in a homeodomain-DNA complex. Biochemistry 34, 14601–14608. - PubMed
-
- Andreeva A, and Murzin AG (2006). Evolution of protein fold in the presence of functional constraints. Curr. Opin. Struct. Biol 16, 399–408. - PubMed
-
- Benach J, Lee I, Edstrom W, Kuzin AP, Chiang Y, Acton TB, Montelione GT, and Hunt JF (2003). The 2.3-Å crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase. J. Biol. Chem 278, 19176–19182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

