A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division
- PMID: 20696759
- PMCID: PMC2952251
- DOI: 10.1074/jbc.M110.142430
A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division
Abstract
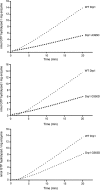
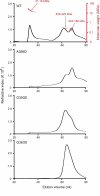
Mitochondria dynamically fuse and divide within cells, and the proper balance of fusion and fission is necessary for normal mitochondrial function, morphology, and distribution. Drp1 is a dynamin-related GTPase required for mitochondrial fission in mammalian cells. It harbors four distinct domains: GTP-binding, middle, insert B, and GTPase effector. A lethal mutation (A395D) within the Drp1 middle domain was reported in a neonate with microcephaly, abnormal brain development, optic atrophy, and lactic acidemia (Waterham, H. R., Koster, J., van Roermund, C. W., Mooyer, P. A., Wanders, R. J., and Leonard, J. V. (2007) N. Engl. J. Med. 356, 1736-1741). Mitochondria within patient-derived fibroblasts were markedly elongated, but the molecular mechanisms underlying these findings were not demonstrated. Because the middle domain is particularly important for the self-assembly of some dynamin superfamily proteins, we tested the hypothesis that this A395D mutation, and two other middle domain mutations (G350D, G363D) were important for Drp1 tetramerization, higher order assembly, and function. Although tetramerization appeared largely intact, each of these mutations compromised higher order assembly and assembly-dependent stimulation of Drp1 GTPase activity. Moreover, mutant Drp1 proteins exhibited impaired localization to mitochondria, indicating that this higher order assembly is important for mitochondrial recruitment, retention, or both. Overexpression of these middle domain mutants markedly inhibited mitochondrial division in cells. Thus, the Drp1 A395D lethal defect likely resulted in impaired higher order assembly of Drp1 at mitochondria, leading to decreased fission, elongated mitochondria, and altered cellular distribution of mitochondria.
Figures






References
-
- Li Z., Okamoto K., Hayashi Y., Sheng M. (2004) Cell 119, 873–887 - PubMed
-
- Arnoult D., Rismanchi N., Grodet A., Roberts R. G., Seeburg D. P., Estaquier J., Sheng M., Blackstone C. (2005) Curr. Biol. 15, 2112–2118 - PubMed
-
- Chan D. C. (2006) Cell 125, 1241–1252 - PubMed
-
- McBride H. M., Neuspiel M., Wasiak S. (2006) Curr. Biol. 16, R551–R560 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

