Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation
- PMID: 20696768
- PMCID: PMC2952260
- DOI: 10.1074/jbc.M110.153817
Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation
Abstract
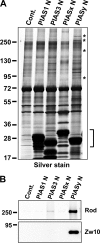
SUMO conjugation of cellular proteins is essential for proper progression of mitosis. PIASy, a SUMO E3 ligase, is required for mitotic SUMOylation of chromosomal proteins, yet the regulatory mechanism behind the PIASy-dependent SUMOylation during mitosis has not been determined. Using a series of truncated PIASy proteins, we have found that the N terminus of PIASy is not required for SUMO modification in vitro but is essential for mitotic SUMOylation in Xenopus egg extracts. We demonstrate that swapping the N terminus of PIASy protein with the corresponding region of other PIAS family members abolishes chromosomal binding and mitotic SUMOylation. We further show that the N-terminal domain of PIASy is sufficient for centromeric localization. We identified that the N-terminal domain of PIASy interacts with the Rod/Zw10 complex, and immunofluorescence further reveals that PIASy colocalizes with Rod/Zw10 in the centromeric region. We show that the Rod/Zw10 complex interacts with the first 47 residues of PIASy which were particularly important for mitotic SUMOylation. Finally, we show that depletion of Rod compromises the centromeric localization of PIASy and SUMO2/3 in mitosis. Together, we demonstrate a fundamental mechanism of PIASy to localize in the centromeric region of chromosome to execute centromeric SUMOylation during mitosis.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

