Mechanism of hypoxia-induced NF-kappaB
- PMID: 20696840
- PMCID: PMC2950552
- DOI: 10.1128/MCB.00409-10
Mechanism of hypoxia-induced NF-kappaB
Abstract
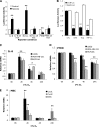
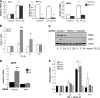
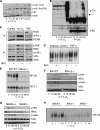
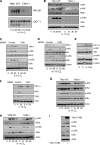
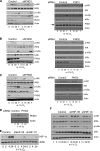
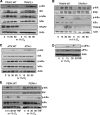
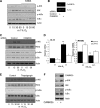
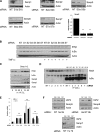
NF-κB activation is a critical component in the transcriptional response to hypoxia. However, the underlying mechanisms that control its activity under these conditions are unknown. Here we report that under hypoxic conditions, IκB kinase (IKK) activity is induced through a calcium/calmodulin-dependent kinase 2 (CaMK2)-dependent pathway distinct from that for other common inducers of NF-κB. This process still requires IKK and the IKK kinase TAK1, like that for inflammatory inducers of NF-κB, but the TAK1-associated proteins TAB1 and TAB2 are not essential. IKK complex activation following hypoxia requires Ubc13 but not the recently identified LUBAC (linear ubiquitin chain assembly complex) ubiquitin conjugation system. In contrast to the action of other NF-κB inducers, IKK-mediated phosphorylation of IκBα does not result in its degradation. We show that this results from IκBα sumoylation by Sumo-2/3 on critical lysine residues, normally required for K-48-linked polyubiquitination. Furthermore, inhibition of specific Sumo proteases is sufficient to release RelA from IκBα and activate NF-κB target genes. These results define a novel pathway regulating NF-κB activation, important to its physiological role in human health and disease.
Figures





References
-
- Adhikari, A., M. Xu, and Z. J. Chen. 2007. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26:3214-3226. - PubMed
-
- Aley, P. K., K. E. Porter, J. P. Boyle, P. J. Kemp, and C. Peers. 2005. Hypoxic modulation of Ca2+ signaling in human venous endothelial cells. Multiple roles for reactive oxygen species. J. Biol. Chem. 280:13349-13354. - PubMed
-
- Bertelsen, M., and A. Sanfridson. 2007. TAB1 modulates IL-1α mediated cytokine secretion but is dispensable for TAK1 activation. Cell. Signal. 19:646-657. - PubMed
-
- Cai, H., D. Liu, and J. G. Garcia. 2008. CaM kinase II-dependent pathophysiological signalling in endothelial cells. Cardiovasc. Res. 77:30-34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
