Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells
- PMID: 20696886
- PMCID: PMC2930588
- DOI: 10.1073/pnas.1006539107
Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells
Abstract
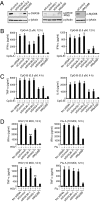
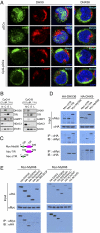
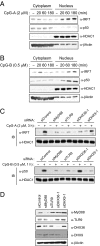
Toll-like receptor 9 (TLR9) senses microbial DNA and triggers type I IFN responses in plasmacytoid dendritic cells (pDCs). Previous studies suggest the presence of myeloid differentiation primary response gene 88 (MyD88)-dependent DNA sensors other than TLR9 in pDCs. Using MS, we investigated C-phosphate-G (CpG)-binding proteins from human pDCs, pDC-cell lines, and interferon regulatory factor 7 (IRF7)-expressing B-cell lines. CpG-A selectively bound the aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicase 36 (DHX36), whereas CpG-B selectively bound DExD/H-box helicase 9 (DHX9). Although the aspartate-glutamate-alanine-histidine box motif (DEAH) domain of DHX36 was essential for CpG-A binding, the domain of unknown function 1605 (DUF1605 domain) of DHX9 was required for CpG-B binding. DHX36 is associated with IFN-alpha production and IRF7 nuclear translocation in response to CpG-A, but DHX9 is important for TNF-alpha and IL-6 production and NF-kappaB activation in response to CpG-B. Knocking down DHX9 or DHX36 significantly reduced the cytokine responses of pDCs to a DNA virus but had no effect on the cytokine responses to an RNA virus. We further showed that both DHX9 and DHX36 are localized within the cytosol and are directly bound to the Toll-interleukin receptor domain of MyD88 via their helicase-associated domain 2 and DUF domains. This study demonstrates that DHX9/DHX36 represent the MyD88-dependent DNA sensors in the cytosol of pDCs and suggests a much broader role for DHX helicases in viral sensing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A DNA-sensing-independent role of a nuclear RNA helicase, DHX9, in stimulation of NF-κB-mediated innate immunity against DNA virus infection.Nucleic Acids Res. 2018 Sep 28;46(17):9011-9026. doi: 10.1093/nar/gky742. Nucleic Acids Res. 2018. PMID: 30137501 Free PMC article.
-
DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells.J Immunol. 2011 Nov 1;187(9):4501-8. doi: 10.4049/jimmunol.1101307. Epub 2011 Sep 28. J Immunol. 2011. PMID: 21957149 Free PMC article.
-
Poxviral protein E3-altered cytokine production reveals that DExD/H-box helicase 9 controls Toll-like receptor-stimulated immune responses.J Biol Chem. 2018 Sep 28;293(39):14989-15001. doi: 10.1074/jbc.RA118.005089. Epub 2018 Aug 15. J Biol Chem. 2018. PMID: 30111593 Free PMC article.
-
DNA Sensing in the Innate Immune Response.Physiology (Bethesda). 2020 Mar 1;35(2):112-124. doi: 10.1152/physiol.00022.2019. Physiology (Bethesda). 2020. PMID: 32027562 Free PMC article. Review.
-
DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication.Biochim Biophys Acta. 2013 Aug;1829(8):854-65. doi: 10.1016/j.bbagrm.2013.03.012. Epub 2013 Apr 6. Biochim Biophys Acta. 2013. PMID: 23567047 Free PMC article. Review.
Cited by
-
Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis.Cancers (Basel). 2024 Jul 11;16(14):2517. doi: 10.3390/cancers16142517. Cancers (Basel). 2024. PMID: 39061157 Free PMC article.
-
BLK positively regulates TLR/IL-1R signaling by catalyzing TOLLIP phosphorylation.J Cell Biol. 2024 Feb 5;223(2):e202302081. doi: 10.1083/jcb.202302081. Epub 2023 Dec 11. J Cell Biol. 2024. PMID: 38078859 Free PMC article.
-
Viral cGAMP nuclease reveals the essential role of DNA sensing in protection against acute lethal virus infection.Sci Adv. 2020 Sep 18;6(38):eabb4565. doi: 10.1126/sciadv.abb4565. Print 2020 Sep. Sci Adv. 2020. PMID: 32948585 Free PMC article.
-
DNA sensor cGAS-mediated immune recognition.Protein Cell. 2016 Nov;7(11):777-791. doi: 10.1007/s13238-016-0320-3. Epub 2016 Sep 30. Protein Cell. 2016. PMID: 27696330 Free PMC article. Review.
-
RNA Helicase A/DHX9 Forms Unique Cytoplasmic Antiviral Granules That Restrict Oncolytic Myxoma Virus Replication in Human Cancer Cells.J Virol. 2021 Jun 24;95(14):e0015121. doi: 10.1128/JVI.00151-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33952639 Free PMC article.
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. - PubMed
-
- Yoneyama M, Fujita T. RIG-I family RNA helicases: Cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. - PubMed
-
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. - PubMed
-
- Schroder K, Muruve DA, Tschopp J. Innate immunity: Cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–R265. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

