Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli
- PMID: 20696893
- PMCID: PMC2932615
- DOI: 10.1073/pnas.1005203107
Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli
Abstract
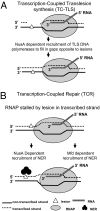
We report observations suggesting that the transcription elongation factor NusA promotes a previously unrecognized class of transcription-coupled repair (TCR) in addition to its previously proposed role in recruiting translesion synthesis (TLS) DNA polymerases to gaps encountered during transcription. Earlier, we reported that NusA physically and genetically interacts with the TLS DNA polymerase DinB (DNA pol IV). We find that Escherichia coli nusA11(ts) mutant strains, at the permissive temperature, are highly sensitive to nitrofurazone (NFZ) and 4-nitroquinolone-1-oxide but not to UV radiation. Gene expression profiling suggests that this sensitivity is unlikely to be due to an indirect effect on gene expression affecting a known DNA repair or damage tolerance pathway. We demonstrate that an N(2)-furfuryl-dG (N(2)-f-dG) lesion, a structural analog of the principal lesion generated by NFZ, blocks transcription by E. coli RNA polymerase (RNAP) when present in the transcribed strand, but not when present in the nontranscribed strand. Our genetic analysis suggests that NusA participates in a nucleotide excision repair (NER)-dependent process to promote NFZ resistance. We provide evidence that transcription plays a role in the repair of NFZ-induced lesions through the isolation of RNAP mutants that display altered ability to survive NFZ exposure. We propose that NusA participates in an alternative class of TCR involved in the identification and removal of a class of lesion, such as the N(2)-f-dG lesion, which are accurately and efficiently bypassed by DinB in addition to recruiting DinB for TLS at gaps encountered by RNAP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
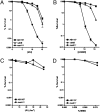
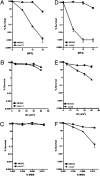
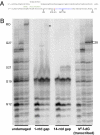
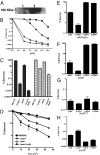
Comment in
-
Linking transcription with DNA repair, damage tolerance, and genome duplication.Proc Natl Acad Sci U S A. 2010 Aug 31;107(35):15314-5. doi: 10.1073/pnas.1010659107. Epub 2010 Aug 23. Proc Natl Acad Sci U S A. 2010. PMID: 20733069 Free PMC article. No abstract available.
References
-
- Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Ed. Washington, DC: Am Soc Microbiol; 2005.
-
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. - PubMed
-
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. - PubMed
-
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. - PubMed
-
- Park JS, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

