Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes
- PMID: 20696906
- PMCID: PMC2930562
- DOI: 10.1073/pnas.1006941107
Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes
Abstract
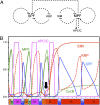
One of the predominant cell-cycle programs found in mature tissues is endoreplication, also known as endoreduplication, that leads to cellular polyploidy. A key question for the understanding of endoreplication cycles is how oscillating levels of cyclin-dependent kinase activity are generated that control repeated rounds of DNA replication. The APC/C performs a pivotal function in the mitotic cell cycle by promoting anaphase and paving the road for a new round of DNA replication. However, using marker lines and plants in which APC/C components are knocked down, we show here that outgrowing and endoreplicating Arabidopsis leaf hairs display no or very little APC/C activity. Instead we find that RBX1-containing Cullin-RING E3 ubiquitin-Ligases (CRLs) are of central importance for the progression through endoreplication cycles; in particular, we have identified CULLIN4 as a major regulator of endoreplication in Arabidopsis trichomes. We have incorporated our findings into a bio-mathematical simulation presenting a robust two-step model of endoreplication control with one type of cyclin-dependent kinase inhibitor function for entry and a CRL-dependent oscillation of cyclin-dependent kinase activity via degradation of a second type of CDK inhibitor during endoreplication cycles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes.Genetics. 2010 May;185(1):257-68. doi: 10.1534/genetics.109.113274. Epub 2010 Mar 1. Genetics. 2010. PMID: 20194967 Free PMC article.
-
Cytokinins promote onset of endoreplication by controlling cell cycle machinery.Plant Signal Behav. 2014;9(8):e29396. doi: 10.4161/psb.29396. Plant Signal Behav. 2014. PMID: 25763620 Free PMC article.
-
The CDK Inhibitor SIAMESE Targets Both CDKA;1 and CDKB1 Complexes to Establish Endoreplication in Trichomes.Plant Physiol. 2020 Sep;184(1):165-175. doi: 10.1104/pp.20.00271. Epub 2020 Jul 21. Plant Physiol. 2020. PMID: 32694133 Free PMC article.
-
Regulation of cullin RING ligases.Annu Rev Plant Biol. 2008;59:467-89. doi: 10.1146/annurev.arplant.58.032806.104011. Annu Rev Plant Biol. 2008. PMID: 18444905 Review.
-
The COP9 signalosome and its role in plant development.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):157-62. doi: 10.1016/j.ejcb.2009.11.021. Epub 2009 Dec 24. Eur J Cell Biol. 2010. PMID: 20036030 Review.
Cited by
-
APRF1 Interactome Reveals HSP90 as a New Player in the Complex That Epigenetically Regulates Flowering Time in Arabidopsis thaliana.Int J Mol Sci. 2024 Jan 21;25(2):1313. doi: 10.3390/ijms25021313. Int J Mol Sci. 2024. PMID: 38279311 Free PMC article.
-
The boundary of the meristematic and elongation zones in roots: endoreduplication precedes rapid cell expansion.Sci Rep. 2013 Oct 14;3:2723. doi: 10.1038/srep02723. Sci Rep. 2013. PMID: 24121463 Free PMC article.
-
Leaf development: a cellular perspective.Front Plant Sci. 2014 Jul 31;5:362. doi: 10.3389/fpls.2014.00362. eCollection 2014. Front Plant Sci. 2014. PMID: 25132838 Free PMC article. Review.
-
Tissue-Specific Control of the Endocycle by the Anaphase Promoting Complex/Cyclosome Inhibitors UVI4 and DEL1.Plant Physiol. 2017 Sep;175(1):303-313. doi: 10.1104/pp.17.00785. Epub 2017 Jul 11. Plant Physiol. 2017. PMID: 28698355 Free PMC article.
-
Effects of the skp1 gene of the SCF complex on lipid metabolism and response to abiotic stress in Chlamydomonas reinhardtii.Front Plant Sci. 2025 Mar 17;16:1527439. doi: 10.3389/fpls.2025.1527439. eCollection 2025. Front Plant Sci. 2025. PMID: 40166727 Free PMC article.
References
-
- Jakoby M, Schnittger A. Cell cycle and differentiation. Curr Opin Plant Biol. 2004;7:661–669. - PubMed
-
- Harashima H, Schnittger A. The integration of cell division, growth and differentiation. Curr Opin Plant Biol. 2010;13:66–74. - PubMed
-
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. - PubMed
-
- Sugimoto-Shirasu K, Roberts K. “Big it up”: Endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6:544–553. - PubMed
-
- Kondorosi E, Roudier F, Gendreau E. Plant cell-size control: Growing by ploidy? Curr Opin Plant Biol. 2000;3:488–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

