Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen
- PMID: 20696907
- PMCID: PMC2930560
- DOI: 10.1073/pnas.1009025107
Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen
Abstract
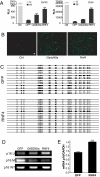
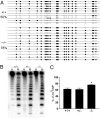
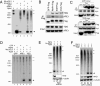
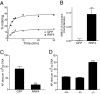
DNA methylation is an important epigenetic modification involved in transcriptional regulation, nuclear organization, development, aging, and disease. Although DNA methyltransferases have been characterized, the mechanisms for DNA demethylation remain poorly understood. Using a cell-based reporter assay, we performed a functional genomics screen to identify genes involved in DNA demethylation. Here we show that RNF4 (RING finger protein 4), a SUMO-dependent ubiquitin E3-ligase previously implicated in maintaining genome stability, plays a key role in active DNA demethylation. RNF4 reactivates methylation-silenced reporters and promotes global DNA demethylation. Rnf4 deficiency is embryonic lethal with higher levels of methylation in genomic DNA. Mechanistic studies show that RNF4 interacts with and requires the base excision repair enzymes TDG and APE1 for active demethylation. This activity appears to occur by enhancing the enzymatic activities that repair DNA G:T mismatches generated from methylcytosine deamination. Collectively, our study reveals a unique function for RNF4, which may serve as a direct link between epigenetic DNA demethylation and DNA repair in mammalian cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
-
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. - PubMed
-
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. - PubMed
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

