Structure and function of the polymerase core of TRAMP, a RNA surveillance complex
- PMID: 20696927
- PMCID: PMC2930566
- DOI: 10.1073/pnas.1003505107
Structure and function of the polymerase core of TRAMP, a RNA surveillance complex
Abstract
The Trf4p/Air2p/Mtr4p polyadenylation (TRAMP) complex recognizes aberrant RNAs in Saccharomyces cerevisiae and targets them for degradation. A TRAMP subcomplex consisting of a noncanonical poly(A) RNA polymerase in the Pol ss superfamily of nucleotidyl transferases, Trf4p, and a zinc knuckle protein, Air2p, mediates initial substrate recognition. Trf4p and related eukaryotic poly(A) and poly(U) polymerases differ from other characterized enzymes in the Pol ss superfamily both in sequence and in the lack of recognizable nucleic acid binding motifs. Here we report, at 2.7-A resolution, the structure of Trf4p in complex with a fragment of Air2p comprising two zinc knuckle motifs. Trf4p consists of a catalytic and central domain similar in fold to those of other noncanonical Pol beta RNA polymerases, and the two zinc knuckle motifs of Air2p interact with the Trf4p central domain. The interaction surface on Trf4p is highly conserved across eukaryotes, providing evidence that the Trf4p/Air2p complex is conserved in higher eukaryotes as well as in yeast and that the TRAMP complex may also function in RNA surveillance in higher eukaryotes. We show that Air2p, and in particular sequences encompassing a zinc knuckle motif near its N terminus, modulate Trf4p activity, and we present data supporting a role for this zinc knuckle in RNA binding. Finally, we show that the RNA 3' end plays a role in substrate recognition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
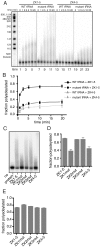
 . Time courses are for Trf4p/Air2pZK1-5 (ZK1-5, lanes 1–12) and Trf4p/Air2pZK4-5 (ZK4-5, lanes 13–24). (B) Plot showing the fraction of
. Time courses are for Trf4p/Air2pZK1-5 (ZK1-5, lanes 1–12) and Trf4p/Air2pZK4-5 (ZK4-5, lanes 13–24). (B) Plot showing the fraction of  adenylated at given time points. The data are from three separate experiments. SEM is indicated. (C) Mutant
adenylated at given time points. The data are from three separate experiments. SEM is indicated. (C) Mutant  was reacted with Trf4p/Air2pZK1-5, Trf4p/Air2pZK4-5, or Trf4p/Air2pZK1-5 but with the three most N-terminal knuckles individually replaced by hexaserine linkers (ZK1mut, ZK2mut, ZK3mut). (D) Quantitation of three experiments as in C. SEM indicated. (E) A5 oligonucleotide is polyadenylated comparably by all Trf4p/Air2p complexes used in C and D.
was reacted with Trf4p/Air2pZK1-5, Trf4p/Air2pZK4-5, or Trf4p/Air2pZK1-5 but with the three most N-terminal knuckles individually replaced by hexaserine linkers (ZK1mut, ZK2mut, ZK3mut). (D) Quantitation of three experiments as in C. SEM indicated. (E) A5 oligonucleotide is polyadenylated comparably by all Trf4p/Air2p complexes used in C and D.


Similar articles
-
RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex.Proc Natl Acad Sci U S A. 2012 May 8;109(19):7292-7. doi: 10.1073/pnas.1201085109. Epub 2012 Apr 24. Proc Natl Acad Sci U S A. 2012. PMID: 22532666 Free PMC article.
-
Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation.Nucleic Acids Res. 2012 Jul;40(12):5679-93. doi: 10.1093/nar/gks223. Epub 2012 Mar 8. Nucleic Acids Res. 2012. PMID: 22402490 Free PMC article.
-
The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex.Cell. 2011 Jun 10;145(6):890-901. doi: 10.1016/j.cell.2011.05.010. Cell. 2011. PMID: 21663793 Free PMC article.
-
Cotranscriptional recruitment of RNA exosome cofactors Rrp47p and Mpp6p and two distinct Trf-Air-Mtr4 polyadenylation (TRAMP) complexes assists the exonuclease Rrp6p in the targeting and degradation of an aberrant messenger ribonucleoprotein particle (mRNP) in yeast.J Biol Chem. 2013 Nov 1;288(44):31816-29. doi: 10.1074/jbc.M113.491290. Epub 2013 Sep 18. J Biol Chem. 2013. PMID: 24047896 Free PMC article.
-
Emerging themes in non-coding RNA quality control.Curr Opin Struct Biol. 2007 Apr;17(2):209-14. doi: 10.1016/j.sbi.2007.03.012. Epub 2007 Mar 28. Curr Opin Struct Biol. 2007. PMID: 17395456 Review.
Cited by
-
Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity.Wiley Interdiscip Rev RNA. 2013 Mar-Apr;4(2):217-31. doi: 10.1002/wrna.1155. Epub 2013 Feb 15. Wiley Interdiscip Rev RNA. 2013. PMID: 23417976 Free PMC article. Review.
-
Purification and Reconstitution of the S. cerevisiae TRAMP and Ski Complexes for Biochemical and Structural Studies.Methods Mol Biol. 2020;2062:491-513. doi: 10.1007/978-1-4939-9822-7_24. Methods Mol Biol. 2020. PMID: 31768992
-
Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing.Genome Res. 2011 Sep;21(9):1478-86. doi: 10.1101/gr.114744.110. Epub 2011 Aug 3. Genome Res. 2011. PMID: 21813626 Free PMC article.
-
Nuclear noncoding RNA surveillance: is the end in sight?Trends Genet. 2012 Jul;28(7):306-13. doi: 10.1016/j.tig.2012.03.005. Epub 2012 Apr 2. Trends Genet. 2012. PMID: 22475369 Free PMC article. Review.
-
Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.Nat Struct Mol Biol. 2012 Aug;19(8):782-787. doi: 10.1038/nsmb.2329. Epub 2012 Jul 1. Nat Struct Mol Biol. 2012. PMID: 22751018 Free PMC article.
References
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. - PubMed
-
- Brosnan CA, Voinnet O. The long and short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. - PubMed
-
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. - PubMed
-
- Anderson JT, Wang X. Nuclear RNA surveillance: No sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

