Dominant spinal muscular atrophy with lower extremity predominance: linkage to 14q32
- PMID: 20697106
- PMCID: PMC2918478
- DOI: 10.1212/WNL.0b013e3181ec800c
Dominant spinal muscular atrophy with lower extremity predominance: linkage to 14q32
Abstract
Objective: Spinal muscular atrophies (SMAs) are hereditary disorders characterized by weakness from degeneration of spinal motor neurons. Although most SMA cases with proximal weakness are recessively inherited, rare families with dominant inheritance have been reported. We aimed to clinically, pathologically, and genetically characterize a large North American family with an autosomal dominant proximal SMA.
Methods: Affected family members underwent clinical and electrophysiologic evaluation. Twenty family members were genotyped on high-density genome-wide SNP arrays and linkage analysis was performed.
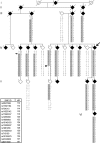
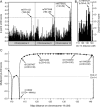
Results: Ten affected individuals (ages 7-58 years) showed prominent quadriceps atrophy, moderate to severe weakness of quadriceps and hip abductors, and milder degrees of weakness in other leg muscles. Upper extremity strength and sensation was normal. Leg weakness was evident from early childhood and was static or very slowly progressive. Electrophysiology and muscle biopsies were consistent with chronic denervation. SNP-based linkage analysis showed a maximum 2-point lod score of 5.10 (theta = 0.00) at rs17679127 on 14q32. A disease-associated haplotype spanning from 114 cM to the 14q telomere was identified. A single recombination narrowed the minimal genomic interval to Chr14: 100,220,765-106,368,585. No segregating copy number variations were found within the disease interval.
Conclusions: We describe a family with an early onset, autosomal dominant, proximal SMA with a distinctive phenotype: symptoms are limited to the legs and there is notable selectivity for the quadriceps. We demonstrate linkage to a 6.1-Mb interval on 14q32 and propose calling this disorder spinal muscular atrophy-lower extremity, dominant.
Figures

References
-
- Pestronk A. Hereditary motor syndromes. Available at: http://neuromuscular.wustl.edu. Accessed November 16, 2009.
-
- Zerres K, Rudnik-Schoneborn S. 93rd ENMC International Workshop: non-5q-spinal muscular atrophies (SMA): clinical picture. Neuromuscul Disord 2003;13:179–183. - PubMed
-
- Grohmann K, Schuelke M, Diers A, et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nature Genet 2001;29:75–77. - PubMed
-
- Christodoulou K, Zamba E, Tsingis M, et al. A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1-p12. Ann Neurol 2000;48:877–884. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
