Refinement of tools for targeted gene expression in Drosophila
- PMID: 20697123
- PMCID: PMC2942869
- DOI: 10.1534/genetics.110.119917
Refinement of tools for targeted gene expression in Drosophila
Abstract
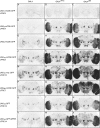
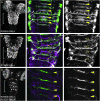
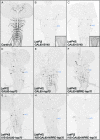
A wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.
Figures


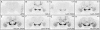





References
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. - PubMed
-
- Ahmad, K., and S. Henikoff, 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104 839–847. - PubMed
-
- Bellen, H. J., C. Wilson and W. J. Gehring, 1990. Dissecting the complexity of the nervous system by enhancer detection. BioEssays 12 199–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

