Transcriptional regulation of Wnt inhibitory factor-1 by Miz-1/c-Myc
- PMID: 20697356
- PMCID: PMC3230129
- DOI: 10.1038/onc.2010.322
Transcriptional regulation of Wnt inhibitory factor-1 by Miz-1/c-Myc
Abstract
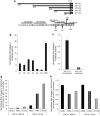
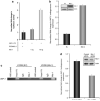
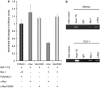
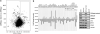
The Wnt signaling pathway is capable of self-regulation through positive and negative feedback mechanisms. For example, the oncoprotein c-Myc, which is upregulated by Wnt signaling activity, participates in a positive feedback loop of canonical Wnt signaling through repression of Wnt antagonists DKK1 and SFRP1. In this study, we investigated the mechanism of Wnt inhibitory factor-1 (WIF-1) silencing. Mapping of CpG island methylation of the WIF-1 promoter reveals regional methylation (-295 to -95 bp from the transcription start site) that correlates with transcriptional silencing. We identified Miz-1 as a direct activator of WIF-1 transcriptional activity, which is found at WIF-1 promoter. In addition, we show that c-Myc contributes to WIF-1 transcriptional repression in a Miz-1-dependent manner. Although the transient repression mediated by Miz-1/c-Myc is independent of de novo methylation, the stable repression by this complex is associated with CpG island methylation of the critical -295 to -95-bp region of the WIF-1 promoter. Importantly, Miz-1 and c-Myc are found at WIF-1 promoter in WIF-1 non-expressing cell lines DLD-1 and 209myc. Transient knockdown or somatic knockout of c-Myc in DLD-1 failed to restore WIF-1 expression suggesting that c-Myc is involved in initiating rather than maintaining WIF-1 epigenetic silencing. In a genome-wide screen, DNAJA4, TGFβ-induced and TRIM59 were repressed by c-Myc overexpression and DNA promoter hypermethylation. Our data reveal novel insights into c-Myc-mediated DNA methylation-dependent transcriptional silencing, a mechanism that might contribute to the dysregulation of Wnt signaling in cancer.
Figures



References
-
- Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcino-genesis. 2006;27:1341–1348. - PubMed
-
- Barr LF, Campbell SE, Diette GB, Gabrielson EW, Kim S, Shim H, et al. c-Myc suppresses the tumorigenicity of lung cancer cells and down-regulates vascular endothelial growth factor expression. Cancer Res. 2000;60:143–149. - PubMed
-
- Benanti JA, Wang ML, Myers HE, Robinson KL, Grandori C, Galloway DA. Epigenetic down-regulation of ARF expression is a selection step in immortalization of human fibroblasts by c-Myc. Mol Cancer Res. 2007;5:1181–1189. - PubMed
-
- Bowen H, Biggs TE, Phillips E, Baker ST, Perry VH, Mann DA, et al. c-Myc represses and Miz-1 activates the murine natural resistance-associated protein 1 promoter. J Biol Chem. 2002;277:34997–35006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

