Plasma proteome response to severe burn injury revealed by 18O-labeled "universal" reference-based quantitative proteomics
- PMID: 20698492
- PMCID: PMC2945297
- DOI: 10.1021/pr1005026
Plasma proteome response to severe burn injury revealed by 18O-labeled "universal" reference-based quantitative proteomics
Abstract
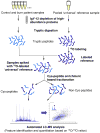
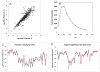
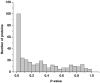
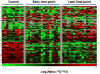
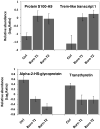
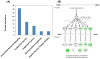
A burn injury represents one of the most severe forms of human trauma and is responsible for significant mortality worldwide. Here, we present the first quantitative proteomics investigation of the blood plasma proteome response to severe burn injury by comparing the plasma protein concentrations of 10 healthy control subjects with those of 15 severe burn patients at two time-points following the injury. The overall analytical strategy for this work integrated immunoaffinity depletion of the 12 most abundant plasma proteins with cysteinyl-peptide enrichment-based fractionation prior to LC-MS analyses of individual patient samples. Incorporation of an 18O-labeled "universal" reference among the sample sets enabled precise relative quantification across samples. In total, 313 plasma proteins confidently identified with two or more unique peptides were quantified. Following statistical analysis, 110 proteins exhibited significant abundance changes in response to the burn injury. The observed changes in protein concentrations suggest significant inflammatory and hypermetabolic response to the injury, which is supported by the fact that many of the identified proteins are associated with acute phase response signaling, the complement system, and coagulation system pathways. The regulation of approximately 35 proteins observed in this study is in agreement with previous results reported for inflammatory or burn response, but approximately 50 potentially novel proteins previously not known to be associated with burn response or inflammation are also found. Elucidating proteins involved in the response to severe burn injury may reveal novel targets for therapeutic interventions as well as potential predictive biomarkers for patient outcomes such as multiple organ failure.
Figures
Similar articles
-
Large-scale multiplexed quantitative discovery proteomics enabled by the use of an (18)O-labeled "universal" reference sample.J Proteome Res. 2009 Jan;8(1):290-9. doi: 10.1021/pr800467r. J Proteome Res. 2009. PMID: 19053531 Free PMC article.
-
Determination of burn patient outcome by large-scale quantitative discovery proteomics.Crit Care Med. 2013 Jun;41(6):1421-34. doi: 10.1097/CCM.0b013e31827c072e. Crit Care Med. 2013. PMID: 23507713 Free PMC article.
-
Production of 18O-single labeled peptide fragments during trypsin digestion of proteins for quantitative proteomics using nanoLC-ESI-MS/MS.J Proteome Res. 2010 Jul 2;9(7):3741-9. doi: 10.1021/pr900865p. J Proteome Res. 2010. PMID: 20496949
-
18O stable isotope labeling in MS-based proteomics.Brief Funct Genomic Proteomic. 2009 Mar;8(2):136-44. doi: 10.1093/bfgp/eln055. Epub 2009 Jan 16. Brief Funct Genomic Proteomic. 2009. PMID: 19151093 Free PMC article. Review.
-
Sample preparation strategies for targeted proteomics via proteotypic peptides in human blood using liquid chromatography tandem mass spectrometry.Proteomics Clin Appl. 2015 Feb;9(1-2):5-16. doi: 10.1002/prca.201400121. Epub 2014 Dec 28. Proteomics Clin Appl. 2015. PMID: 25418444 Review.
Cited by
-
18Oxygen Substituted Nucleosides Combined with Proton Beam Therapy: Therapeutic Transmutation In Vitro.Int J Part Ther. 2021 Mar 19;7(4):11-18. doi: 10.14338/IJPT-D-20-00036.1. eCollection 2021 Spring. Int J Part Ther. 2021. PMID: 33829069 Free PMC article.
-
Meta-analysis of peptides to detect protein significance.Stat Interface. 2020;13(4):465-474. doi: 10.4310/sii.2020.v13.n4.a4. Epub 2020 Jul 31. Stat Interface. 2020. PMID: 34055134 Free PMC article.
-
Lymphatic Vessel Network Structure and Physiology.Compr Physiol. 2018 Dec 13;9(1):207-299. doi: 10.1002/cphy.c180015. Compr Physiol. 2018. PMID: 30549020 Free PMC article. Review.
-
Mass spectrometry in cancer biomarker research: a case for immunodepletion of abundant blood-derived proteins from clinical tissue specimens.Biomark Med. 2014;8(2):269-86. doi: 10.2217/bmm.13.101. Biomark Med. 2014. PMID: 24521024 Free PMC article.
-
Genomics of injury: The Glue Grant experience.J Trauma Acute Care Surg. 2015 Apr;78(4):671-86. doi: 10.1097/TA.0000000000000568. J Trauma Acute Care Surg. 2015. PMID: 25742245 Free PMC article. No abstract available.
References
-
- Atiyeh BS, Costagliola M, Hayek SN. Burn prevention mechanisms and outcomes: pitfalls, failures and successes. Burns. 2009;35 (2):181–93. - PubMed
-
- Atiyeh BS, Gunn SW, Dibo SA. Metabolic implications of severe burn injuries and their management: a systematic review of the literature. World J Surg. 2008;32 (8):1857–69. - PubMed
-
- Herndon DN, Tompkins RG. Support of the metabolic response to burn injury. Lancet. 2004;363 (9424):1895–902. - PubMed
-
- Matsuda N, Hattori Y. Systemic inflammatory response syndrome (SIRS): molecular pathophysiology and gene therapy. J Pharmacol Sci. 2006;101 (3):189–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

