baySeq: empirical Bayesian methods for identifying differential expression in sequence count data
- PMID: 20698981
- PMCID: PMC2928208
- DOI: 10.1186/1471-2105-11-422
baySeq: empirical Bayesian methods for identifying differential expression in sequence count data
Abstract
Background: High throughput sequencing has become an important technology for studying expression levels in many types of genomic, and particularly transcriptomic, data. One key way of analysing such data is to look for elements of the data which display particular patterns of differential expression in order to take these forward for further analysis and validation.
Results: We propose a framework for defining patterns of differential expression and develop a novel algorithm, baySeq, which uses an empirical Bayes approach to detect these patterns of differential expression within a set of sequencing samples. The method assumes a negative binomial distribution for the data and derives an empirically determined prior distribution from the entire dataset. We examine the performance of the method on real and simulated data.
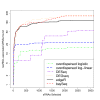
Conclusions: Our method performs at least as well, and often better, than existing methods for analyses of pairwise differential expression in both real and simulated data. When we compare methods for the analysis of data from experimental designs involving multiple sample groups, our method again shows substantial gains in performance. We believe that this approach thus represents an important step forward for the analysis of count data from sequencing experiments.
Figures





References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
-
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

