Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes
- PMID: 20699272
- PMCID: PMC2952877
- DOI: 10.1093/nar/gkq673
Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes
Abstract
An array of nano-channels was fabricated from silicon based semiconductor materials to stretch long, native dsDNA. Here we present a labeling scheme in which it is possible to identify the location of specific sequences along the stretched DNA molecules. The scheme proceeds by first using the strand displacement activity of the Vent (exo-) polymerase to generate single strand flaps on nicked dsDNA. These single strand flaps are hybridized with sequence specific fluorophore-labeled probes. Subsequent imaging of the DNA molecules inside a nano-channel array device allows for quantitative identification of the location of probes. The highly efficient DNA hybridization on the ss-DNA flaps is an excellent method to identify the sequence motifs of dsDNA as it gives us unique ability to control the length of the probe sequence and thus the frequency of hybridization sites on the DNA. We have also shown that this technique can be extended to a multi color labeling scheme by using different dye labeled probes or by combining with a DNA- polymerase-mediated incorporation of fluorophore-labeled nucleotides on nicking sites. Thus this labeling chemistry in conjunction with the nano-channel platform can be a powerful tool to solve complex structural variations in DNA which is of importance for both research and clinical diagnostics of genetic diseases.
Figures
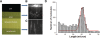




References
-
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. - PubMed
-
- Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 2002;11:1987–1995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

