Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis
- PMID: 20699384
- PMCID: PMC2930549
- DOI: 10.1073/pnas.1006432107
Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis
Abstract
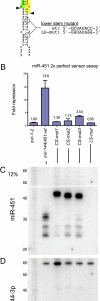
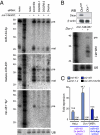
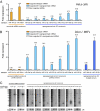
Canonical animal microRNAs (miRNAs) are generated by sequential cleavage of precursor substrates by the Drosha and Dicer RNase III enzymes. Several variant pathways exploit other RNA metabolic activities to generate functional miRNAs. However, all of these pathways culminate in Dicer cleavage, suggesting that this is a unifying feature of miRNA biogenesis. Here, we show that maturation of miR-451, a functional miRNA that is perfectly conserved among vertebrates, is independent of Dicer. Instead, structure-function and knockdown studies indicate that Drosha generates a short pre-mir-451 hairpin that is directly cleaved by Ago2 and followed by resection of its 3' terminus. We provide stringent evidence for this model by showing that Dicer knockout cells can generate mature miR-451 but not other miRNAs, whereas Ago2 knockout cells reconstituted with wild-type Ago2, but not Slicer-deficient Ago2, can process miR-451. Finally, we show that the mir-451 backbone is amenable to reprogramming, permitting vector-driven expression of diverse functional miRNAs in the absence of Dicer. Beyond the demonstration of an alternative strategy to direct gene silencing, these observations open the way for transgenic rescue of Dicer conditional knockouts.
Figures

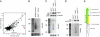
Similar articles
-
Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates.Cell Cycle. 2010 Nov 15;9(22):4455-60. doi: 10.4161/cc.9.22.13958. Epub 2010 Nov 15. Cell Cycle. 2010. PMID: 21088485 Free PMC article.
-
Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis.Mol Cell. 2020 Apr 16;78(2):317-328.e6. doi: 10.1016/j.molcel.2020.02.020. Epub 2020 Mar 18. Mol Cell. 2020. PMID: 32191872 Free PMC article.
-
Functional parameters of Dicer-independent microRNA biogenesis.RNA. 2012 May;18(5):945-57. doi: 10.1261/rna.032938.112. Epub 2012 Mar 29. RNA. 2012. PMID: 22461413 Free PMC article.
-
Dicer-independent processing of small RNA duplexes: mechanistic insights and applications.Nucleic Acids Res. 2017 Oct 13;45(18):10369-10379. doi: 10.1093/nar/gkx779. Nucleic Acids Res. 2017. PMID: 28977573 Free PMC article. Review.
-
Versatile microRNA biogenesis in animals and their viruses.RNA Biol. 2014;11(6):673-81. doi: 10.4161/rna.28985. Epub 2014 May 7. RNA Biol. 2014. PMID: 24823351 Free PMC article. Review.
Cited by
-
RNase III-independent microRNA biogenesis in mammalian cells.RNA. 2012 Dec;18(12):2166-73. doi: 10.1261/rna.036194.112. Epub 2012 Oct 24. RNA. 2012. PMID: 23097423 Free PMC article.
-
Loop-miRs: active microRNAs generated from single-stranded loop regions.Nucleic Acids Res. 2013 May 1;41(10):5503-12. doi: 10.1093/nar/gkt251. Epub 2013 Apr 10. Nucleic Acids Res. 2013. PMID: 23580554 Free PMC article.
-
Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):E4007-16. doi: 10.1073/pnas.1510476112. Epub 2015 Jul 13. Proc Natl Acad Sci U S A. 2015. PMID: 26170322 Free PMC article.
-
MicroRNA biogenesis: regulating the regulators.Crit Rev Biochem Mol Biol. 2013 Jan-Feb;48(1):51-68. doi: 10.3109/10409238.2012.738643. Epub 2012 Nov 19. Crit Rev Biochem Mol Biol. 2013. PMID: 23163351 Free PMC article. Review.
-
Development of Novel Small Hairpin RNAs That do not Require Processing by Dicer or AGO2.Mol Ther. 2016 Aug;24(7):1278-89. doi: 10.1038/mt.2016.81. Epub 2016 Apr 25. Mol Ther. 2016. PMID: 27109632 Free PMC article.
References
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. - PubMed
-
- Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

