A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature
- PMID: 20699401
- PMCID: PMC2949010
- DOI: 10.1104/pp.110.157610
A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature
Abstract
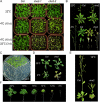
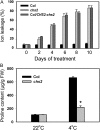
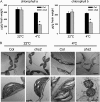
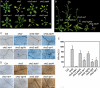
How plants adapt to low temperature is not well understood. To identify components involved in low-temperature signaling, we characterized the previously isolated chilling-sensitive2 mutant (chs2) of Arabidopsis (Arabidopsis thaliana). This mutant grew normally at 22°C but showed phenotypes similar to activation of defense responses when shifted to temperatures below 16°C. These phenotypes include yellowish and collapsed leaves, increased electrolyte leakage, up-regulation of PATHOGENESIS RELATED genes, and accumulation of excess hydrogen peroxide and salicylic acid (SA). Moreover, the chs2 mutant was seedling lethal when germinated at or shifted for more than 3 d to low temperatures of 4°C to 12°C. Map-based cloning revealed that a single amino acid substitution occurred in the TIR-NB-LRR (for Toll/Interleukin-1 receptor- nucleotide-binding Leucine-rich repeat)-type resistance (R) protein RPP4 (for Recognition of Peronospora parasitica4), which causes a deregulation of the R protein in a temperature-dependent manner. The chs2 mutation led to an increase in the mutated RPP4 mRNA transcript, activation of defense responses, and an induction of cell death at low temperatures. In addition, a chs2 intragenic suppressor, in which the mutation occurs in the conserved NB domain, abolished defense responses at lower temperatures. Genetic analyses of chs2 in combination with known SA pathway and immune signaling mutants indicate that the chs2-conferred temperature sensitivity requires ENHANCED DISEASE SUSCEPTIBILITY1, REQUIRED FOR Mla12 RESISTANCE, and SUPPRESSOR OF G2 ALLELE OF skp1 but does not require PHYTOALEXIN DEFICIENT4, NONEXPRESSOR OF PR GENES1, or SA. This study reveals that an activated TIR-NB-LRR protein has a large impact on temperature sensitivity in plant growth and survival.
Figures




Similar articles
-
Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses.New Phytol. 2014 Jun;202(4):1320-1334. doi: 10.1111/nph.12760. Epub 2014 Mar 11. New Phytol. 2014. PMID: 24611624
-
A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis.Plant J. 2013 Aug;75(4):553-65. doi: 10.1111/tpj.12232. Epub 2013 Jun 7. Plant J. 2013. PMID: 23651299
-
Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation.Plant Physiol. 2013 Jul;162(3):1694-705. doi: 10.1104/pp.113.214551. Epub 2013 May 20. Plant Physiol. 2013. PMID: 23690534 Free PMC article.
-
A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis.Plant J. 2010 Jul;63(2):283-296. doi: 10.1111/j.1365-313X.2010.04241.x. Epub 2010 Apr 28. Plant J. 2010. PMID: 20444230
-
A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature.Plant J. 2013 Aug;75(4):539-52. doi: 10.1111/tpj.12219. Epub 2013 May 22. Plant J. 2013. PMID: 23617639
Cited by
-
Identification of multiple novel genetic mechanisms that regulate chilling tolerance in Arabidopsis.Front Plant Sci. 2023 Jan 12;13:1094462. doi: 10.3389/fpls.2022.1094462. eCollection 2022. Front Plant Sci. 2023. PMID: 36714785 Free PMC article.
-
Cosuppression of RBCS3B in Arabidopsis leads to severe photoinhibition caused by ROS accumulation.Plant Cell Rep. 2014 Jul;33(7):1091-108. doi: 10.1007/s00299-014-1597-4. Epub 2014 Mar 30. Plant Cell Rep. 2014. PMID: 24682522
-
Natural variation in temperature-modulated immunity uncovers transcription factor bHLH059 as a thermoresponsive regulator in Arabidopsis thaliana.PLoS Genet. 2021 Jan 25;17(1):e1009290. doi: 10.1371/journal.pgen.1009290. eCollection 2021 Jan. PLoS Genet. 2021. PMID: 33493201 Free PMC article.
-
A new eye on NLR proteins: focused on clarity or diffused by complexity?Curr Opin Immunol. 2012 Feb;24(1):41-50. doi: 10.1016/j.coi.2011.12.006. Epub 2012 Feb 3. Curr Opin Immunol. 2012. PMID: 22305607 Free PMC article. Review.
-
BRASSINOSTEROID-INSENSITIVE2 Negatively Regulates the Stability of Transcription Factor ICE1 in Response to Cold Stress in Arabidopsis.Plant Cell. 2019 Nov;31(11):2682-2696. doi: 10.1105/tpc.19.00058. Epub 2019 Aug 13. Plant Cell. 2019. PMID: 31409630 Free PMC article.
References
-
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 - PubMed
-
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

