A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete
- PMID: 20700437
- PMCID: PMC2916851
- DOI: 10.1371/journal.pgen.1001052
A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete
Abstract
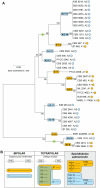
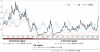
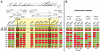
In fungi, sexual identity is determined by specialized genomic regions called MAT loci which are the equivalent to sex chromosomes in some animals and plants. Usually, only two sexes or mating types exist, which are determined by two alternate sets of genes (or alleles) at the MAT locus (bipolar system). However, in the phylum Basidiomycota, a unique tetrapolar system emerged in which four different mating types are generated per meiosis. This occurs because two functionally distinct molecular recognition systems, each encoded by one MAT region, constrain the selection of sexual partners. Heterozygosity at both MAT regions is a pre-requisite for mating in both bipolar and tetrapolar basidiomycetes. Tetrapolar mating behaviour results from the absence of genetic linkage between the two regions bringing forth up to thousands of mating types. The subphylum Pucciniomycotina, an early diverged lineage of basidiomycetes encompassing important plant pathogens such as the rusts and saprobes like Rhodosporidium and Sporidiobolus, has been so far poorly explored concerning the content and organization of MAT loci. Here we show that the red yeast Sporidiobolus salmonicolor has a mating system unlike any previously described because occasional disruptions of the genetic cohesion of the bipolar MAT locus originate new mating types. We confirmed that mating is normally bipolar and that heterozygosity at both MAT regions is required for mating. However, a laboratory cross showed that meiotic recombination may occur within the bipolar MAT locus, explaining tetrapolar features like increased allele number and evolution rates of some MAT genes. This pseudo-bipolar system deviates from the classical bipolar-tetrapolar paradigm and, to our knowledge, has never been observed before. We propose a model for MAT evolution in the Basidiomycota in which the pseudo-bipolar system may represent a hitherto unforeseen gradual form of transition from an ancestral tetrapolar system to bipolarity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Fungal Sex: The Basidiomycota.Microbiol Spectr. 2017 Jun;5(3):10.1128/microbiolspec.funk-0046-2016. doi: 10.1128/microbiolspec.FUNK-0046-2016. Microbiol Spectr. 2017. PMID: 28597825 Free PMC article. Review.
-
Evolution of Mating Systems in Basidiomycetes and the Genetic Architecture Underlying Mating-Type Determination in the Yeast Leucosporidium scottii.Genetics. 2015 Sep;201(1):75-89. doi: 10.1534/genetics.115.177717. Epub 2015 Jul 14. Genetics. 2015. PMID: 26178967 Free PMC article.
-
Convergent evolution of linked mating-type loci in basidiomycete fungi.PLoS Genet. 2019 Sep 6;15(9):e1008365. doi: 10.1371/journal.pgen.1008365. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31490920 Free PMC article.
-
Identification of mating type genes in the bipolar basidiomycetous yeast Rhodosporidium toruloides: first insight into the MAT locus structure of the Sporidiobolales.Eukaryot Cell. 2008 Jun;7(6):1053-61. doi: 10.1128/EC.00025-08. Epub 2008 Apr 11. Eukaryot Cell. 2008. PMID: 18408057 Free PMC article.
-
Orchestration of sexual reproduction and virulence by the fungal mating-type locus.Curr Opin Microbiol. 2008 Dec;11(6):517-24. doi: 10.1016/j.mib.2008.09.014. Epub 2008 Nov 5. Curr Opin Microbiol. 2008. PMID: 18935978 Free PMC article. Review.
Cited by
-
The cacao pathogen Moniliophthora roreri (Marasmiaceae) possesses biallelic A and B mating loci but reproduces clonally.Heredity (Edinb). 2016 Jun;116(6):491-501. doi: 10.1038/hdy.2016.5. Epub 2016 Mar 2. Heredity (Edinb). 2016. PMID: 26932308 Free PMC article.
-
Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex.PLoS Genet. 2012;8(2):e1002528. doi: 10.1371/journal.pgen.1002528. Epub 2012 Feb 16. PLoS Genet. 2012. PMID: 22359516 Free PMC article.
-
Fungal Sex: The Basidiomycota.Microbiol Spectr. 2017 Jun;5(3):10.1128/microbiolspec.funk-0046-2016. doi: 10.1128/microbiolspec.FUNK-0046-2016. Microbiol Spectr. 2017. PMID: 28597825 Free PMC article. Review.
-
Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis.mBio. 2013 Jan 22;4(1):e00572-12. doi: 10.1128/mBio.00572-12. mBio. 2013. PMID: 23341551 Free PMC article.
-
Comparative genomics of the mating-type loci of the mushroom Flammulina velutipes reveals widespread synteny and recent inversions.PLoS One. 2011;6(7):e22249. doi: 10.1371/journal.pone.0022249. Epub 2011 Jul 20. PLoS One. 2011. PMID: 21799803 Free PMC article.
References
-
- Fraser JA, Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr Opin Genet Dev. 2005;15:645–651. - PubMed
-
- Rodriguez-Carres M, Findley K, Sun S, Dietrich FS, Heitman J. Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS ONE. 2010;5:e9620. doi: 10.1371/journal.pone.0009620. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

