Lysine120 interactions with p53 response elements can allosterically direct p53 organization
- PMID: 20700496
- PMCID: PMC2916859
- DOI: 10.1371/journal.pcbi.1000878
Lysine120 interactions with p53 response elements can allosterically direct p53 organization
Abstract
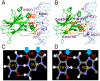
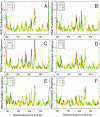
p53 can serve as a paradigm in studies aiming to figure out how allosteric perturbations in transcription factors (TFs) triggered by small changes in DNA response element (RE) sequences, can spell selectivity in co-factor recruitment. p53-REs are 20-base pair (bp) DNA segments specifying diverse functions. They may be located near the transcription start sites or thousands of bps away in the genome. Their number has been estimated to be in the thousands, and they all share a common motif. A key question is then how does the p53 protein recognize a particular p53-RE sequence among all the similar ones? Here, representative p53-REs regulating diverse functions including cell cycle arrest, DNA repair, and apoptosis were simulated in explicit solvent. Among the major interactions between p53 and its REs involving Lys120, Arg280 and Arg248, the bps interacting with Lys120 vary while the interacting partners of other residues are less so. We observe that each p53-RE quarter site sequence has a unique pattern of interactions with p53 Lys120. The allosteric, DNA sequence-induced conformational and dynamic changes of the altered Lys120 interactions are amplified by the perturbation of other p53-DNA interactions. The combined subtle RE sequence-specific allosteric effects propagate in the p53 and in the DNA. The resulting amplified allosteric effects far away are reflected in changes in the overall p53 organization and in the p53 surface topology and residue fluctuations which play key roles in selective co-factor recruitment. As such, these observations suggest how similar p53-RE sequences can spell the preferred co-factor binding, which is the key to the selective gene transactivation and consequently different functional effects.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Cooperativity dominates the genomic organization of p53-response elements: a mechanistic view.PLoS Comput Biol. 2009 Jul;5(7):e1000448. doi: 10.1371/journal.pcbi.1000448. Epub 2009 Jul 24. PLoS Comput Biol. 2009. PMID: 19629163 Free PMC article.
-
Structural Basis for p53 Lys120-Acetylation-Dependent DNA-Binding Mode.J Mol Biol. 2016 Jul 31;428(15):3013-25. doi: 10.1016/j.jmb.2016.06.009. Epub 2016 Jun 23. J Mol Biol. 2016. PMID: 27338200
-
Distinct mechanisms control genome recognition by p53 at its target genes linked to different cell fates.Nat Commun. 2021 Jan 20;12(1):484. doi: 10.1038/s41467-020-20783-z. Nat Commun. 2021. PMID: 33473123 Free PMC article.
-
Transcriptional regulation by p53: one protein, many possibilities.Cell Death Differ. 2006 Jun;13(6):951-61. doi: 10.1038/sj.cdd.4401916. Cell Death Differ. 2006. PMID: 16575405 Review.
-
The Rich World of p53 DNA Binding Targets: The Role of DNA Structure.Int J Mol Sci. 2019 Nov 9;20(22):5605. doi: 10.3390/ijms20225605. Int J Mol Sci. 2019. PMID: 31717504 Free PMC article. Review.
Cited by
-
DNA-binding protects p53 from interactions with cofactors involved in transcription-independent functions.Nucleic Acids Res. 2016 Nov 2;44(19):9096-9109. doi: 10.1093/nar/gkw770. Epub 2016 Sep 6. Nucleic Acids Res. 2016. PMID: 27604871 Free PMC article.
-
Raf/MEK/ERK Signaling Pathway Is Involved in the Inhibition of Glioma Cell Proliferation and Invasion in the Ketogenic Microenvironment.Curr Med Sci. 2023 Aug;43(4):759-767. doi: 10.1007/s11596-023-2724-7. Epub 2023 Jul 27. Curr Med Sci. 2023. PMID: 37498407
-
The role of response elements organization in transcription factor selectivity: the IFN-β enhanceosome example.PLoS Comput Biol. 2011 Jun;7(6):e1002077. doi: 10.1371/journal.pcbi.1002077. Epub 2011 Jun 16. PLoS Comput Biol. 2011. PMID: 21698143 Free PMC article.
-
Molecular dynamics of the full-length p53 monomer.Cell Cycle. 2013 Sep 15;12(18):3098-108. doi: 10.4161/cc.26162. Epub 2013 Sep 5. Cell Cycle. 2013. PMID: 23974096 Free PMC article.
-
Roles of computational modelling in understanding p53 structure, biology, and its therapeutic targeting.J Mol Cell Biol. 2019 Apr 1;11(4):306-316. doi: 10.1093/jmcb/mjz009. J Mol Cell Biol. 2019. PMID: 30726928 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

