Transcription regulation of the Escherichia coli pcnB gene coding for poly(A) polymerase I: roles of ppGpp, DksA and sigma factors
- PMID: 20700605
- PMCID: PMC2939334
- DOI: 10.1007/s00438-010-0567-y
Transcription regulation of the Escherichia coli pcnB gene coding for poly(A) polymerase I: roles of ppGpp, DksA and sigma factors
Abstract
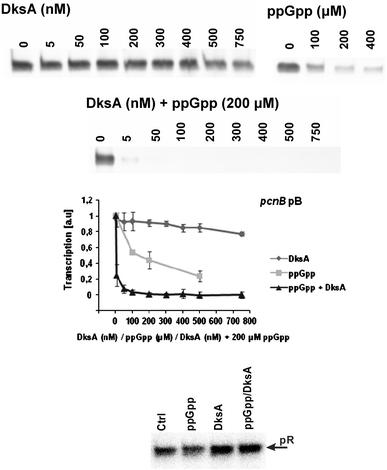
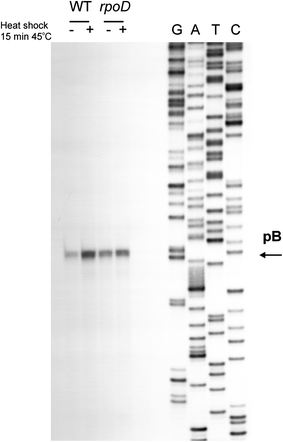
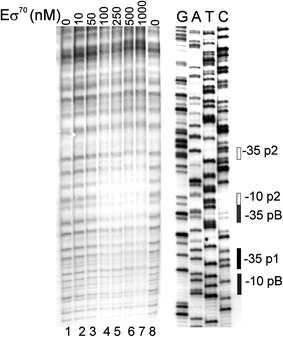
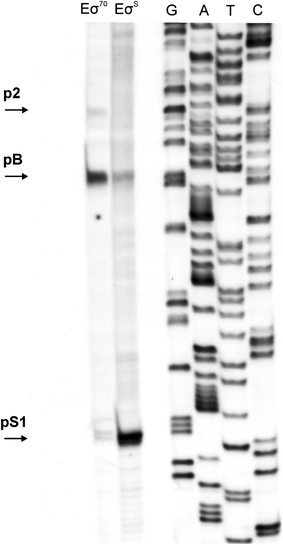
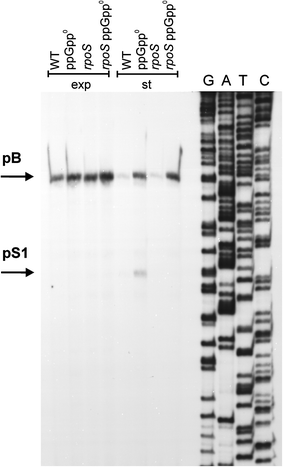
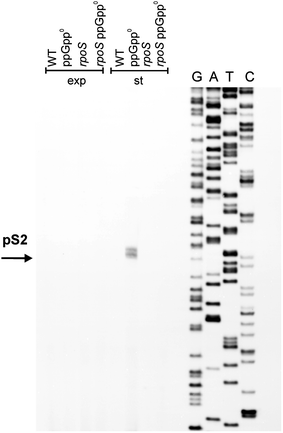
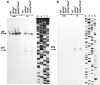
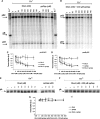
Poly(A) polymerase I (PAP I), encoded by the pcnB gene, is a major enzyme responsible for RNA polyadenylation in Escherichia coli, a process involved in the global control of gene expression in this bacterium through influencing the rate of transcript degradation. Recent studies have suggested a complicated regulation of pcnB expression, including a complex promoter region, a control at the level of translation initiation and dependence on bacterial growth rate. In this report, studies on transcription regulation of the pcnB gene are described. Results of in vivo and in vitro experiments indicated that (a) there are three σ(70)-dependent (p1, pB, and p2) and two σ(S)-dependent (pS1 and pS2) promoters of the pcnB gene, (b) guanosine tetraphosphate (ppGpp) and DksA directly inhibit transcription from pB, pS1 and pS2, and (c) pB activity is drastically impaired at the stationary phase of growth. These results indicate that regulation of the pcnB gene transcription is a complex process, which involves several factors acting to ensure precise control of PAP I production. Moreover, inhibition of activities of pS1 and pS2 by ppGpp and DksA suggests that regulation of transcription from promoters requiring alternative σ factors by these effectors of the stringent response might occur according to both passive and active models.
Figures


Similar articles
-
DksA and ppGpp Regulate the σS Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP.J Bacteriol. 2017 Dec 20;200(2):e00463-17. doi: 10.1128/JB.00463-17. Print 2018 Jan 15. J Bacteriol. 2017. PMID: 29061665 Free PMC article.
-
Transcription start sites in the promoter region of the Escherichia coli pcnB (plasmid copy number) gene coding for poly(A) polymerase I.Plasmid. 2006 Mar;55(2):169-72. doi: 10.1016/j.plasmid.2005.10.002. Epub 2005 Dec 2. Plasmid. 2006. PMID: 16330100
-
The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription.Mol Microbiol. 2006 May;60(3):749-64. doi: 10.1111/j.1365-2958.2006.05129.x. Mol Microbiol. 2006. PMID: 16629675
-
CbrAB-dependent regulation of pcnB, a poly(A) polymerase gene involved in polyadenylation of RNA in Pseudomonas fluorescens.Environ Microbiol. 2010 Jun;12(6):1674-83. doi: 10.1111/j.1462-2920.2010.02228.x. Epub 2010 May 7. Environ Microbiol. 2010. PMID: 20482591
-
Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity.FEMS Microbiol Rev. 2010 Sep;34(5):646-57. doi: 10.1111/j.1574-6976.2010.00223.x. Epub 2010 Apr 14. FEMS Microbiol Rev. 2010. PMID: 20491934 Review.
Cited by
-
High-Throughput Screening of a Promoter Library Reveals New Persister Mechanisms in Escherichia Coli.Microbiol Spectr. 2022 Feb 23;10(1):e0225321. doi: 10.1128/spectrum.02253-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196813 Free PMC article.
-
Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100.J Bacteriol. 2011 Apr;193(7):1745-56. doi: 10.1128/JB.01343-10. Epub 2011 Jan 28. J Bacteriol. 2011. PMID: 21278289 Free PMC article.
-
RNA polyadenylation and its consequences in prokaryotes.Philos Trans R Soc Lond B Biol Sci. 2018 Nov 5;373(1762):20180166. doi: 10.1098/rstb.2018.0166. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30397102 Free PMC article. Review.
-
Tyrosine phosphorylation controlled poly(A) polymerase I activity regulates general stress response in bacteria.Life Sci Alliance. 2022 Dec 19;6(3):e202101148. doi: 10.26508/lsa.202101148. Print 2023 Mar. Life Sci Alliance. 2022. PMID: 36535710 Free PMC article.
-
Transgenesis of mammalian PABP reveals mRNA polyadenylation as a general stress response mechanism in bacteria.iScience. 2021 Sep 11;24(10):103119. doi: 10.1016/j.isci.2021.103119. eCollection 2021 Oct 22. iScience. 2021. PMID: 34646982 Free PMC article.
References
-
- August JT, Ortiz PJ, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962;237:3786–3793. - PubMed
-
- Baracchini E, Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J Biol Chem. 1988;263:2597–2602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

