Ranked retrieval of Computational Biology models
- PMID: 20701772
- PMCID: PMC2936397
- DOI: 10.1186/1471-2105-11-423
Ranked retrieval of Computational Biology models
Abstract
Background: The study of biological systems demands computational support. If targeting a biological problem, the reuse of existing computational models can save time and effort. Deciding for potentially suitable models, however, becomes more challenging with the increasing number of computational models available, and even more when considering the models' growing complexity. Firstly, among a set of potential model candidates it is difficult to decide for the model that best suits ones needs. Secondly, it is hard to grasp the nature of an unknown model listed in a search result set, and to judge how well it fits for the particular problem one has in mind.
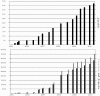
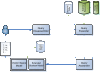
Results: Here we present an improved search approach for computational models of biological processes. It is based on existing retrieval and ranking methods from Information Retrieval. The approach incorporates annotations suggested by MIRIAM, and additional meta-information. It is now part of the search engine of BioModels Database, a standard repository for computational models.
Conclusions: The introduced concept and implementation are, to our knowledge, the first application of Information Retrieval techniques on model search in Computational Systems Biology. Using the example of BioModels Database, it was shown that the approach is feasible and extends the current possibilities to search for relevant models. The advantages of our system over existing solutions are that we incorporate a rich set of meta-information, and that we provide the user with a relevance ranking of the models found for a query. Better search capabilities in model databases are expected to have a positive effect on the reuse of existing models.
Figures


References
-
- Klipp E, Liebermeister W, Wierling C, Kowald A, Lehrach H, Herwig R. Systems biology: a textbook. Wiley-VCH; 2009.
-
- Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, Snoep JL, Hucka M, Novére NL, Laibe C. BioModels Database: An enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. - DOI - PMC - PubMed
-
- Liebermeister W. Validity and combination of biochemical models. Proceedings of 3rd International ESCEC Workshop on Experimental Standard Conditions on Enzyme Characterizations. 2008.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

