Programming cells: towards an automated 'Genetic Compiler'
- PMID: 20702081
- PMCID: PMC2950163
- DOI: 10.1016/j.copbio.2010.07.005
Programming cells: towards an automated 'Genetic Compiler'
Abstract
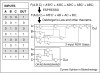
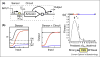
One of the visions of synthetic biology is to be able to program cells using a language that is similar to that used to program computers or robotics. For large genetic programs, keeping track of the DNA on the level of nucleotides becomes tedious and error prone, requiring a new generation of computer-aided design (CAD) software. To push the size of projects, it is important to abstract the designer from the process of part selection and optimization. The vision is to specify genetic programs in a higher-level language, which a genetic compiler could automatically convert into a DNA sequence. Steps towards this goal include: defining the semantics of the higher-level language, algorithms to select and assemble parts, and biophysical methods to link DNA sequence to function. These will be coupled to graphic design interfaces and simulation packages to aid in the prediction of program dynamics, optimize genes, and scan projects for errors.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures



References
-
- Ham TS, Lee SK, Keasling JD, Arkin AP. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotech Bioeng. 2006;94:1–4. - PubMed
-
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotech. 2006;17:548–557. - PubMed
-
- Salis H, Kaznessis YN. Computer-aided design of modular protein devices: Boolean AND gene activation. Physical Biology. 2006;3:395–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

