Selection systems based on dominant-negative transcription factors for precise genetic engineering
- PMID: 20702421
- PMCID: PMC2965260
- DOI: 10.1093/nar/gkq708
Selection systems based on dominant-negative transcription factors for precise genetic engineering
Abstract
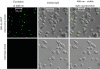
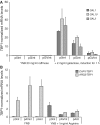
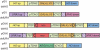
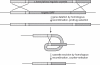
Diverse tools are available for performing genetic modifications of microorganisms. However, new methods still need to be developed for performing precise genomic engineering without introducing any undesirable side-alteration. Indeed for functional analyses of genomic elements, as well as for some industrial applications, only the desired mutation should be introduced at the locus considered. This article describes a new approach fulfilling these requirements, based on the use of selection systems consisting in truncated genes encoding dominant-negative transcription factors. We have demonstrated dominant-negative effects mediated by truncated Gal4p and Arg81p proteins in Saccharomyces cerevisiae, interfering with galactose and arginine metabolic pathways, respectively. These genes can be used as positive and negative markers, since they provoke both growth inhibition on substrates and resistance to specific drugs. These selection markers have been successfully used for precisely deleting HO and URA3 in wild yeasts. This genetic engineering approach could be extended to other microorganisms.
Figures

References
-
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. - PubMed
-
- Giaver G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. - PubMed
-
- Johnston M, Riles L, Hegemann JH. Gene disruption. Methods Enzymol. 2000;350:290–315. - PubMed
-
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

