Elucidation of the conformational free energy landscape in H.pylori LuxS and its implications to catalysis
- PMID: 20704697
- PMCID: PMC2929236
- DOI: 10.1186/1472-6807-10-27
Elucidation of the conformational free energy landscape in H.pylori LuxS and its implications to catalysis
Abstract
Background: One of the major challenges in understanding enzyme catalysis is to identify the different conformations and their populations at detailed molecular level in response to ligand binding/environment. A detail description of the ligand induced conformational changes provides meaningful insights into the mechanism of action of enzymes and thus its function.
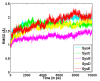
Results: In this study, we have explored the ligand induced conformational changes in H.pylori LuxS and the associated mechanistic features. LuxS, a dimeric protein, produces the precursor (4,5-dihydroxy-2,3-pentanedione) for autoinducer-2 production which is a signalling molecule for bacterial quorum sensing. We have performed molecular dynamics simulations on H.pylori LuxS in its various ligand bound forms and analyzed the simulation trajectories using various techniques including the structure network analysis, free energy evaluation and water dynamics at the active site. The results bring out the mechanistic details such as co-operativity and asymmetry between the two subunits, subtle changes in the conformation as a response to the binding of active and inactive forms of ligands and the population distribution of different conformations in equilibrium. These investigations have enabled us to probe the free energy landscape and identify the corresponding conformations in terms of network parameters. In addition, we have also elucidated the variations in the dynamics of water co-ordination to the Zn2+ ion in LuxS and its relation to the rigidity at the active sites.
Conclusions: In this article, we provide details of a novel method for the identification of conformational changes in the different ligand bound states of the protein, evaluation of ligand-induced free energy changes and the biological relevance of our results in the context of LuxS structure-function. The methodology outlined here is highly generalized to illuminate the linkage between structure and function in any protein of known structure.
Figures

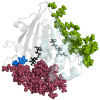




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

